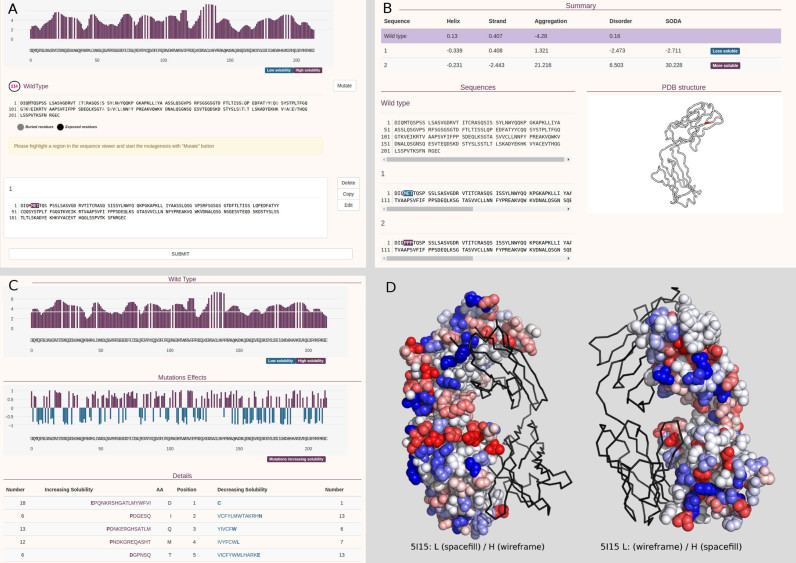
Figure 1.
Sample SODA mutation and result pages. (A) The mutation input page is returned when the user chooses ‘mutation mode’. It allows to create multiple instances of mutations/deletions/insertions. (B) The ‘mutation mode’ result reports changes of the protein solubility upon mutation. When a PDB is provided as input the mutated region is highlighted in the structure. (C) The ‘full protein mode’ provides the solubility profile (first plot) and the propensity of increasing or decreasing solubility for all sequence positions (second plot and table). (D) The human germline antibody IGHV1-69/IGKV1-39 (PDB code 5i15) is shown alternatively as wireframe and space fill between light (L) and heavy (H) chain. For each position, the probability of increasing (red) or decreasing (blue) solubility upon mutation is mapped on the structure. On the left, the light (L) and heavy (H) chains are shown as wireframe and space fill respectively, on the right the same protein with opposite chain visualization mode is provided.