Abstract
Background: Zika virus (ZIKV) was first discovered in East Africa in 1947. ZIKV has caused microcephaly in the Americas, but it is not known whether ZIKV is a cause of microcephaly in East Africa.
Methods: We used surveillance data from 11,061 live births at Kilifi County Hospital in coastal Kenya between January 2012 and October 2016 to identify microcephaly cases and conducted a nested case-control study to determine risk factors for microcephaly. Gestational age at birth was estimated based on antenatal ultrasound scanning (‘Scanned cohort’) or last menstrual period (‘LMP cohort’, including births ≥37 weeks’ gestation only). Controls were newborns with head circumference Z scores between >-2 and ≤2 SD that were compared to microcephaly cases in relation to ZIKV exposure and other maternal and newborn factors.
Results: Of the 11,061 newborns, 214 (1.9%, 95%CI 1.69, 2.21) had microcephaly. Microcephaly prevalence was 1.0% (95%CI 0.64, 1.70, n=1529) and 2.1% (95%CI 1.81, 2.38, n=9532) in the scanned and LMP cohorts, respectively. After excluding babies <2500 g (n=1199) in the LMP cohort the prevalence was 1.1% (95%CI 0.93, 1.39). Microcephaly showed an association with being born small for gestational age (p<0.001) but not with ZIKV neutralising antibodies (p=0.6) or anti-ZIKV NS1 IgM response (p=0.9). No samples had a ZIKV neutralising antibody titre that was at least fourfold higher than the corresponding dengue virus (DENV) titre. No ZIKV or other flavivirus RNA was detected in cord blood from cases or controls.
Conclusions: Microcephaly was prevalent in coastal Kenya, but does not appear to be related to ZIKV exposure; the ZIKV response observed in our study population was largely due to cross-reactive responses to DENV or other related flaviviruses. Further research into potential causes and the clinical consequences of microcephaly in this population is urgently needed.
Keywords: Congenital microcephaly, Flavivirus, Zika virus
Introduction
The recent Zika virus (ZIKV) epidemic in the Americas has focused attention on microcephaly as a major complication of in-utero infection and a cause of neurodisability in newborns 1. Very little is known about the burden of microcephaly in Africa and, though ZIKV was first discovered in East Africa 2 and the Aedes mosquito vector for ZIKV is plentiful, it is not known whether ZIKV is a cause of microcephaly in the region. A cross-sectional survey in 1966-68 found high (52%) ZIKV antibody seroprevalence among children and adults in coastal Kenya 3, though antibody cross-reactivity between ZIKV and other flaviviruses in circulation such as dengue virus (DENV) and West Nile virus (WNV) makes the interpretation of these data difficult. Several major flavivirus outbreaks have since occurred in the country 4– 6, with various serosurveys indicating ongoing flavivirus exposure 7, 8. Notably, high flavivirus antibody seroprevalence was reported amongst pregnant women sampled in 2002-03 in coastal Kenya but the association with birth outcomes was not determined 9.
We previously initiated a perinatal and maternal health research programme in coastal Kenya to identify risk factors for: 1) severe morbidity and mortality in mothers and newborns 10 and 2) preterm and small for gestational age (SGA) births in the INTERBIO-21 st Study 11. As part of these two studies, we took head circumference measurements and demographic and anthropometric data allowing an estimation of: 1) the prevalence of microcephaly in coastal Kenya; 2) its association with maternal and newborn factors, and 3) its association with flavivirus exposure.
Methods
Study population and data collection
This was a population-based, observational, cohort study undertaken at Kilifi County Hospital (KCH) between January 2012 and October 2016. KCH is a rural public county hospital providing comprehensive obstetric care annually to approximately 5,000 women living along the Kenyan coast. All women completed a standardised admission record as part of two studies: an ongoing clinical surveillance study assessing risk factors for severe morbidity and mortality in mothers and newborns 10 and the INTERBIO-21 st Study 11. This included socio-demographic information, clinical history including antenatal clinic attendance, clinical findings on admission, delivery details, and maternal and newborn anthropometry. Gestational age was determined either by calculating the difference between the date of delivery and the date of the last reported menstrual period (LMP), including only births ≥37 weeks’ gestation (“LMP cohort”); or by a pregnancy dating ultrasound scan done ≤24 weeks’ gestation for a subset of participants enrolled in the INTERBIO-21 st Study 11, which included preterm and term births, referred to hereafter as the “scanned cohort”.
All newborns had anthropometric measurements (i.e. head circumference, weight and length) taken within 48 hours of birth by nurses and fieldworkers trained as part of the INTERBIO-21 st Study, which included quarterly refresher training and continual quality control. Anthropometry for the scanned cohort was done in duplicate by two different fieldworkers, and discrepancies resolved by a third measurement. 11 Maternal blood for routine and research samples was collected on admission, and umbilical cord blood was collected at delivery. Maternal and cord blood samples were processed, and plasma stored at -80ºC, within 24 hours of collection. All mothers provided written informed consent for use of their biological samples and clinical data. The studies were approved by the Kenya Medical Research Institute (KEMRI) Scientific and Ethics Review Unit (KEMRI SERU # 3296 and 1778).
Laboratory procedures
Viral RNA detection. For qRT-PCR detection of ZIKV and other flaviviruses, viral RNA was isolated from cord plasma using the QIAamp® Viral RNA kit (Qiagen) according to manufacturer’s instructions. Samples were then screened for ZIKV and other flavivirus RNA using the QuantiFast RT-PCR kit (Qiagen) and published pan-flavivirus (Flavi allS, Flavi all AS2, Flavi all AS4 and Flavi all probe 3 mix) 12 and ZIKV-specific primers and probes, Bonn E and Bonn NS1 13, on an ABI 7500 Real Time PCR system. Sequences for all primers can be found in the indicated references. The PCR cycling conditions for the pan-flavivirus assay 12 were: 50°C for 20 minutes, 95°C for 15 minutes, followed by 45 cycles comprising 95°C for 15 seconds and 60°C for 1 minute. For ZIKV Bonn E and Bonn NS1 13 the conditions were: 50°C for 20 minutes, 95°C for 15 minutes, followed by 45 cycles comprising 95°C for 15 seconds and 58°C for 1 minute. A cycle threshold value of <40 was used to define positives for all three assays. RNA isolated from ZIKV MR766 strain and a range of other flaviviruses (DENV, WNV and Yellow Fever virus) cultured in Vero E6 cells were used as positive controls, and the PCR mastermix without template used as a negative control in these assays.
FRNT 90 assay. Cord plasma were screened for antibodies to ZIKV using a ZIKV focus reduction neutralisation test (FRNT 90) and an in-house IgM ELISA assay against ZIKV NS1 antigen strain MR766. For the FRNT 90 assay, heat-inactivated cord plasma samples were diluted to 1:20 in 100 µl Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal calf serum (D10), mixed with an equal volume of D10 containing approximately 100 focus-forming units of ZIKV MR766 strain, and incubated for 1 hour at 37ºC. The virus-plasma mixture was then overlaid onto 96-well flat-bottomed plates containing Vero E6 monolayers at 90% confluency for virus adsorption at 37ºC, 5% CO 2 for 24 hours. The virus-plasma mixture was aspirated from the wells, 100 µl of D10 was added and the plates incubated for a further 24 hours at 37ºC, 5% CO 2. Immunostaining was then used to detect virus infection. Briefly, cells were gently washed with phosphate buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 10 minutes and permeabilized with permeabilization buffer (0.5% Triton X-100 in PBS) for 30 minutes. The plates were blocked in Blocker™ Casein (ThermoFisher) before addition of 0.5 µg/ml of the anti-flavivirus E protein monoclonal antibody 4G2 (Native Antigen, UK, Cat. No AbFLAVENV-4G2) in permeabilization buffer for a 2-hour incubation at 37°C. Following a further series of washes, plates were incubated with a 1:1000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody (Abcam, Cat. No. ab6789) in permeabilization buffer for 1 hour at 37°C, and colour development of foci done by addition of 3,3'-diaminobenzidine (Sigma) substrate for 10 minutes at room temperature. The plates were finally washed, air dried and foci counted using an AID ELISpot reader. Plasma samples that resulted in at least 90% reduction in foci relative to wells incubated with virus only were considered flavivirus seropositive. Antibody titres against the ZIKV MR766 strain, and against a local DENV-2 isolate obtained from a patient in coastal Kenya, were then estimated for seropositive samples using the FRNT 90 assay on twofold serial plasma dilutions. FRNT 90 antibody titres were calculated using the Reed and Muench method 14.
ZIKV NS1 IgM ELISA. For the IgM assay, 96-well flat-bottomed plates were first coated with 1 µg/ml of the ZIKV NS1 antigen (Native Antigen, UK, Cat. No. ZIKV-NS1) at room temperature overnight, then washed in wash buffer (0.05% Tween in PBS) and blocked with Blocker™ Casein (ThermoFisher) for 1 hour. Cord plasma were diluted 1:400 in Blocker™ Casein, added to plates in duplicate and incubated for 2 hours at room temperature. After a further series of washes, a 1:5000 dilution of HRP-conjugated goat anti-human IgM antibody (KPL, Cat. No. 074-1003) in wash buffer was added to plates, incubated for 1 hour, washed and OPD substrate (Sigma) added for colour development for 15 minutes. Plates were read on a Biotek ELISA reader at a wavelength of 492 nm and optical density values for each sample acquired for analysis. Plasma from coastal Kenya residents with previous PCR-confirmed DENV infection 15 were used as positive controls, while plasma from two European individuals and a pool of cord plasma from 10 neonates without detectable responses to the ZIKV NS1 antigen were used as negative controls. IgM ratios, defined as the ratio between mean sample OD and the mean OD of the negative controls, were then obtained and seropositivity defined as an IgM ratio of >3 as done by others 16.
Statistical analysis
Based on World Health Organization recommendations 14, microcephaly (cases) was defined as a birth head circumference (HC) Z score < -3 SD from the mean for gestational age and sex using INTERGROWTH-21 st (IG21) newborn size reference charts for births <33 weeks’ gestation 15, and standards for births ≥33 weeks’ gestation 16. Controls were defined as newborns with HC Z scores between > -2 and ≤ 2 SD. Univariable logistic regression models were used to estimate associations between microcephaly and maternal or newborn variables hypothesised to be putative risk factors (22 variables tested; Table 1). A nominal two-sided p value was calculated as <0.002 (i.e. 0.05 divided by 22 for the number of covariates) following Bonferroni adjustment for multiple testing. Variables reaching the nominal p<0.002 were then included in a multivariable logistic regression model and their adjusted association with microcephaly estimated. All logistic regression analyses were performed using pooled data from the scanned and LMP cohorts, respectively. To assess the distribution of variables between the two cohorts stratified analyses were performed using χ 2 tests (for categorical variables) and Mann-Whitney U-tests (for continuous variables). All analyses were carried out in Stata™ version 15 with two-sided p-values reported.
Table 1. Association between maternal and newborn co-factors and microcephaly.
The total number and prevalence of cases in the final case-control dataset, stratified by categories of neonatal and maternal variables, are shown. Data from the scanned and LMP cohorts are pooled in these analyses, but cohort-specific frequencies are shown in Table 3. Crude odds ratios (OR), 95% confidence intervals (CI) and P values from univariable logistic regression models estimating associations with microcephaly with each variable in turn are shown. The reference population in each of the models is assigned a value of 1. The total number of newborns included in each analysis varies due to missing data for some variables. *Systolic or diastolic blood pressure >140 or >90 mmHg, respectively.
| Covariate | Categories | n/N cases (%) | Crude OR (95% CI) | P value |
|---|---|---|---|---|
| Newborn factors | ||||
| Sex | Male | 69/4794 (1.4%) | 1 | |
| Female | 42/4303 (1.0%) | 0.67 (0.46, 0.99) | 0.05 | |
| Small for gestational age (SGA) | Normal | 61/7331 (0.8%) | 1 | |
| SGA | 50/1766 (2.8%) | 3.47 (2.38, 5.07) | <0.001 | |
| Type of birth | Singleton | 106/8879 (1.2%) | 1 | |
| Multifetal | 5/218 (2.3%) | 1.9 (0.80, 4.75) | 0.14 | |
| Year of birth | 2012 | 17/1381 (1.2%) | 1 | |
| 2013 | 28/1499 (1.9%) | 1.53 (0.83, 2.80) | 0.17 | |
| 2014 | 23/2379 (1.0%) | 0.78 (0.42, 1.47) | 0.45 | |
| 2015 | 23/2043 (1.1%) | 0.91 (0.49, 1.72) | 0.78 | |
| 2016 | 20/1795 (1.1%) | 0.90 (0.47, 1.73) | 0.76 | |
| Season | January – March | 24/2245 (1.1%) | 1 | |
| April – June | 41/2742 (1.5%) | 1.40 (0.85, 2.33) | 0.19 | |
| July – September | 22/2240 (1.0%) | 0.92 (0.51, 1.64) | 0.77 | |
| October – December | 24/1870 (1.3%) | 1.20 (0.68, 2.13) | 0.52 | |
| Maternal factors | ||||
| Sociodemographic factors | ||||
| Maternal age | <20 years | 17/1339 (1.3%) | 1 | |
| 20 to 35 years | 87/6966 (1.2%) | 0.98 (0.58, 1.66) | 0.95 | |
| >35 years | 7/783 (0.9%) | 0.70 (0.29, 1.70) | 0.43 | |
| Marital status | Married | 99/8329 (1.2%) | 1 | |
| Unmarried | 10/665 (1.5%) | 1.27 (0.66, 2.44) | 0.47 | |
| Education level | Secondary or more | 21/2627 (0.8%) | 1 | |
| Primary school | 73/5253 (1.4%) | 1.75 (1.07, 2.85) | 0.02 | |
| None | 13/1010 (1.3) | 1.62 (0.81, 3.24) | 0.17 | |
| Residence | Other | 70/5893 (1.2%) | 1 | |
| Kilifi township | 41/3176 (1.3%) | 1.09 (0.74, 1.60) | 0.67 | |
| Type of house | Stone wall | 45/4702 (1.0%) | 1 | |
| Mud wall | 63/4328 (1.5%) | 1.53 (1.04, 2.25) | 0.03 | |
| Obstetric history | ||||
| Parity | Primigravida | 41/3302 (1.2%) | 1 | |
| Multigravida | 70/5738 (1.2%) | 0.98 (0.67, 1.45) | 0.93 | |
| Antenatal care attendance | ≥4 visits | 69/5887 (1.2%) | 1 | |
| 0 to 3 visits | 42/3434 (1.3%) | 1.05 (0.72, 1.55) | 0.79 | |
| Medication during pregnancy | ||||
| Folic acid supplements | Yes | 102/8292 (1.2%) | 1 | |
| No | 9/795 (1.1%) | 0.92 (0.46, 1.82) | 0.81 | |
| Malaria prophylaxis | ≥3 doses | 61/5166 (1.2%) | 1 | |
| 1 to 2 doses | 47/3070 (1.5%) | 1.30 (0.89, 1.91) | 0.18 | |
| None | 3/689 (0.4%) | 0.37 (0.11, 1.17) | 0.09 | |
| Tetanus vaccination | Yes | 95/7706 (1.2%) | 1 | |
| No | 13/1137 (1.1%) | 0.93 (0.52, 1.66) | 0.80 | |
| Maternal co-morbidities and infections | ||||
| Mid-upper arm circumference
(MUAC) |
Normal (23–30cm) | 76/6490 (1.2%) | 1 | |
| Low (<23cm) | 13/990 (1.3%) | 1.12 (0.62, 2.03) | 0.70 | |
| High (≥30cm) | 9/997 (0.9%) | 0.77 (0.38, 1.54) | 0.46 | |
| Hypertension in pregnancy * | No | 92/7623(1.2%) | 1 | |
| Yes | 9/540 (1.7%) | 1.39 (0.70, 2.77) | 0.35 | |
| HIV status | Negative | 107/8605 (1.2%) | 1 | |
| Positive | 5/365 (1.1%) | 0.88 (0.32, 2.40) | 0.80 | |
| Maternal anaemia | No | 21/2321 (0.9%) | 1 | |
| Yes | 66/5436 (1.2%) | 1.35 (0.82, 2.20) | 0.24 | |
| VDRL (syphilis test) | Negative | 95/8014 (1.2%) | 1 | |
| Positive | 1/45 (2.2%) | 1.89 (0.26, 13.89) | 0.53 | |
| Other risk exposures | ||||
| Substance use | No | 108/8855 (1.2%) | 1 | |
| Yes | 3/232 (1.3%) | 1.06 (0.33, 3.37) | 0.92 | |
| Contact with cattle | No | 101/8225 (1.2%) | 1 | |
| Yes | 7/463 (1.5%) | 1.23 (0.57, 2.67) | 0.59 | |
Results
Prevalence of microcephaly
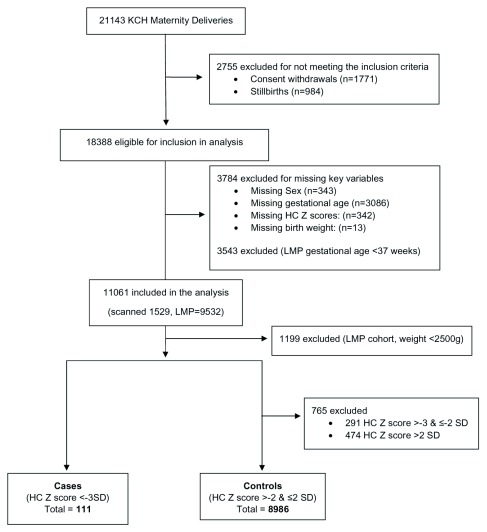
Between January 2012 and October 2016 there were 21,143 births at KCH. We excluded stillbirths (n=984), consent withdrawals (n=1771), births with missing key variables (sex, gestational age, HC Z scores and birth weight; n=3784) and preterm newborns for the LMP cohort only (n=3543). We included 11061 live births in the main analysis ( Figure 1). The mean gestational age of newborns in the scanned cohort was 38.6 weeks (95% CI 38.44, 38.66) and 39.3 weeks (95% CI 39.26, 39.32) for newborns in the LMP cohort.
Figure 1. Study participants flow diagram.
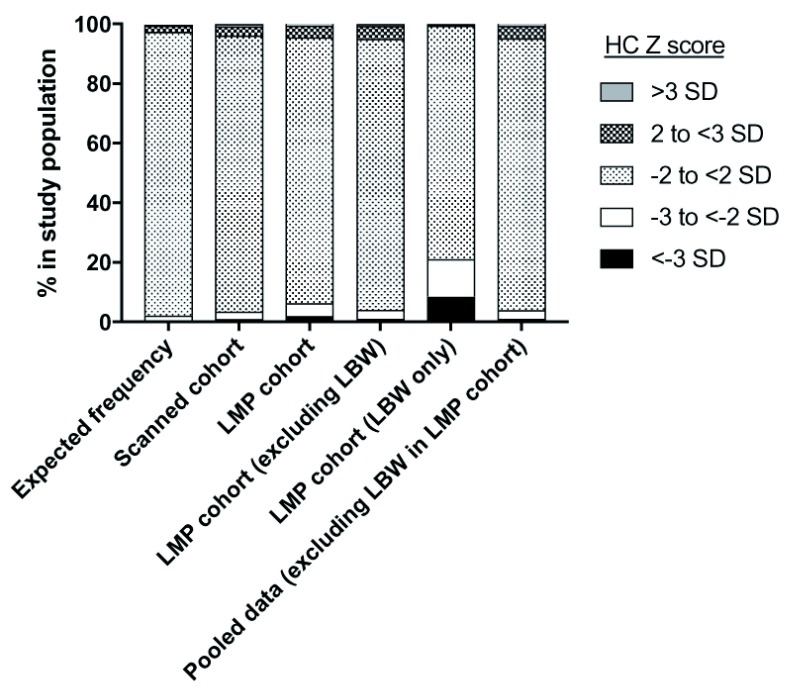
There was an excess frequency of births with HC Z scores below -3 SD in the study population when compared to the expected normal distribution ( Figure 2). However, the observed frequency of births with HC Z scores between -3 and <-2 SD was similar to the expected normal distribution ( Figure 2).
Figure 2. Distribution of head circumference (HC) Z scores in the study population.
The distribution of HC Z scores in the study population, measured as described in the Methods section, are shown. For comparison, the expected frequencies in a normal distribution are shown. For the LMP cohort distributions are shown for the full cohort, after exclusion of newborns with low birth weight (LBW; <2500 g), or for LBW newborns only. Distributions for the final analysis dataset (‘Pooled data [excluding LBW in LMP cohort]) are shown for comparison.
A total of 16 (1.0%) of the 1529 newborns in the scanned cohort, and 198 (2.1%) of the 9532 newborns in the LMP cohort had a HC Z score <-3 SD. We hypothesised that the higher case prevalence in the LMP cohort was due to the known inaccuracy of LMP in estimating gestational age as compared to ultrasound scans, leading to preterm births being classified as term, in turn resulting in a lower Z score than would have been assigned had the gestational age been known accurately 14. Indeed, among 1399 births (1253 term, 146 preterm) in the scanned cohort (used as gold standard) that also had corresponding LMP gestation ages, LMP gestation would have misclassified 9.3% (117/1253) of the term births as preterm births, and 39.7% (58/146) of preterms as term births. Therefore, to enhance the specificity of our case definition for associations, we excluded newborns with low birth weight (<2500 g, as per WHO guidelines) from the analysis of the LMP cohort. When this exclusion was applied the prevalence of microcephaly in the LMP cohort was 1.1% (95% CI 0.93, 1.39) among newborns weighing ≥2500 g (n=8333) and 8.6% (95% CI 7.13, 10.32) among those weighing <2500 g (n=1199), a difference that was statistically significant (χ 2=286.05, p<0.001; Fig. 2). In contrast, low birth weight showed no association with microcephaly in the scanned cohort (χ 2=0.40, p=0.5). All further analyses on the LMP cohort were therefore restricted to newborns weighing ≥2500 g at birth, which when pooled with the scanned cohort gave a case prevalence of 1.1% (95% CI 0.93, 1.35).
Associations between microcephaly and maternal and newborn factors
To identify potential risk factors for microcephaly, we used a nested case-control approach whereby cases (n=111) were compared to controls (newborns with HC Z score > -2 and ≤ 2 SD, n=8986) with respect to various maternal and newborn variables by logistic regression ( Table 1 and Table 3). A strong association was observed between microcephaly and being born small for gestational age (SGA), defined as birth weight <10 th centile for gestational age and sex on IG21 charts (OR=3.47, 95% CI 2.38, 5.07, p<0.001). Maternal nutritional status, anaemia, HIV status, parity, receipt of interventions provided in the antenatal clinic and all other newborn and maternal factors tested showed no significant association with microcephaly.
Associations between microcephaly and flavivirus exposure
We used qRT-PCR and serological assays to investigate whether flavivirus exposure was associated with microcephaly in our dataset. To test for recent exposure to flavivirus, we measured IgM antibody responses against ZIKV NS1 antigen in cord plasma from 94 cases with available samples and 864 controls matched by year of birth. Overall IgM seropositivity against ZIKV NS1 was 2.4% (95% CI 1.60, 3.59), though this was strongly confounded by cross-reactive responses to DENV since sera from 25 patients from coastal Kenya with lab-confirmed dengue infection 17, 18 were all seropositive on the ZIKV NS1 IgM assay. No association was evident between the ZIKV NS1 IgM response and microcephaly ( Table 2). Furthermore, no ZIKV or other flavivirus RNA could be detected in cord plasma from the 94 cases or from a random selection of controls (n=471).
Table 2. Associations between flavivirus serology and microcephaly.
Prevalence of anti-ZIKV antibody responses as measured by FRNT 90 assay and IgM ELISA is shown for cases and controls. Odds ratios, 95% confidence intervals and p value for the association with microcephaly are shown.
| Seropositivity
n/N (%) |
Crude OR (95% CI) | P | ||
|---|---|---|---|---|
| ZIKV NS1 IgM assay | Controls | 21/864 (2.4%) | 1 | |
| Cases | 2/94 (2.1%) | 0.87 (0.20, 3.78) | 0.86 | |
| ZIKV FRNT90 assay | Controls | 61/755 (8.1%) | 1 | |
| Cases | 7/71 (9.7%) | 1.24 (0.55, 2.83) | 0.60 |
Table 3. Stratified analyses estimating associations between microcephaly and maternal and newborn co-factors in each cohort.
Univariate analyses assessing the relationship between maternal and newborn factors and microcephaly in each cohort are shown. For each cohort the frequency of cases is shown, and χ 2 test used for all analyses. When all variables that were statistically significant at p<0.05 in any of the cohorts (indicated by *) were included in a multivariable logistic regression model, only SGA maintained an association with microcephaly (adjusted OR=3.41, 95% CI 2.30, 5.06, p<0.001).
| LMP cohort | Scanned cohort | ||||
|---|---|---|---|---|---|
| Covariate | Categories | n/N cases (%) | P value | n/N cases (%) | P value |
| A) Newborn factors | |||||
| Sex | Male | 58/4047 (1.4%) | 0.10 | 11/747 (1.5%) | 0.18 |
| Female | 37/3618 (1.0%) | 5/685 (0.7%) | |||
| Small for gestational age (SGA) * | Normal | 51/6184 (0.8%) | <0.001 | 10/1147 (0.9%) | 0.08 |
| SGA | 44/1481 (3.0%) | 6/285 (2.1%) | |||
| Type of birth * | Singleton | 92/7472 (1.2%) | 0.69 | 14/1393 (1.0%) | 0.001 |
| Multifetal | 3/193 (1.5%) | 2/25 (8.0%) | |||
| Year of birth | 2012 | 17/1312 (1.3%) | 0.05 | 0/69 (0) | 0.75 |
| 2013 | 25/1225 (2.0%) | 3/274 (1.1%) | |||
| 2014 | 14/1712 (0.8%) | 9/667 (1.3%) | |||
| 2015 | 19/1621 (1.2%) | 4/422 (0.9%) | |||
| 2016 | 20/1795 (1.1%) | No data | |||
| &Season | January - March | 21/1835 (1.1%) | 0.33 | 3/410 (0.7%) | 0.18 |
| April - June | 34/2379 (1.4%) | 7/363 (1.9%) | |||
| July - September | 17/1899 (0.9%) | 5/341 (1.5%) | |||
| October - December | 23/1552 (1.5%) | 1/318 (0.3%) | |||
| B) Maternal factors | |||||
| Sociodemographic factors | |||||
| Maternal age | <20 years | 14/1164 (1.2%) | 0.49 | 3/175 (1.7%) | 0.59 |
| 20 to 35 years | 76/5835 (1.3%) | 11/1131 (1.0%) | |||
| >35 years | 5/659 (0.7%) | 2/124 (1.6%) | |||
| Marital status * | Married | 87/6999 (1.2%) | 0.68 | 12/1330 (0.9%) | 0.002 |
| Unmarried | 6/575 (1.2%) | 4/90 (4.4%) | |||
| Education level | Secondary or more | 20/2221 (0.9%) | 0.22 | 1/406 (0.2%) | 0.15 |
| Primary school | 62/4443 (1.4%) | 11/810 (1.4%) | |||
| None | 10/832 (1.2%) | 3/178 (1.7%) | |||
| Residence | Other | 60/5293 (1.1%) | 0.19 | 10/600 (1.7%) | 0.09 |
| Kilifi township | 35/2346 (1.5%) | 6/830 (0.7%) | |||
| Type of house * | Stone wall | 36/3902 (0.9%) | 0.02 | 9/800 (1.1%) | 0.99 |
| Mud wall | 56/3708 (1.5%) | 7/620 (1.1%) | |||
| Obstetric history | |||||
| Parity | Primigravida | 39/2814 (1.4%) | 0.40 | 2/488 (0.4%) | 0.06 |
| Multigravida | 56/4812 (1.2%) | 14/926 (1.5%) | |||
| Antenatal care | |||||
| Antenatal care attendance | ≥4 visits | 57/4501 (1.3%) | 0.87 | 12/1221 (1.0%) | 0.22 |
| 0 to 3 visits | 38/3102 (1.2%) | 4/203 (2.0%) | |||
| Medication during pregnancy | |||||
| Folic acid supplements | Yes | 86/6947 (1.2%) | 0.95 | 16/1345 (1.2%) | 0.32 |
| No | 9/713 (1.3%) | 0/82 (0) | |||
| Malaria prophylaxis | ≥3 doses | 53/4106 (1.3%) | 0.07 | 8/1060 (0.7%) | 0.05 |
| 1 to 2 doses | 40/2785 (1.4%) | 7/285 (2.5%) | |||
| None | 2/632 (0.3%) | 1/57 (1.7%) | |||
| Tetanus vaccination | Yes | 79/6456 (1.2%) | 0.82 | 16/1250 (1.3%) | 0.17 |
| No | 13/994 (1.3%) | 0/143 (0) | |||
| Maternal co-morbidities and infections | |||||
| Mid-upper arm circumference (MUAC) | Normal (23-30cm) | 65/5417 (1.2%) | 0.85 | 11/1073 (1.0%) | 0.22 |
| Low (<23cm) | 9/822 (1.1%) | 4/168 (2.4%) | |||
| High (≥30cm) | 8/812 (1.0%) | 1/185 (0.5%) | |||
| Hypertension in pregnancy | No | 77/6296 (1.2%) | 0.31 | 15/1327 (1.1%) | 0.996 |
| Yes | 8/452 (1.8%) | 1/88 (1.1%) | |||
| HIV status | Negative | 91/7277 (1.1%) | 0.78 | 16/1328 (1.2%) | 0.30 |
| Positive | 4/278 (1.4%) | 0/87 (0) | |||
| Maternal anaemia | No | 19/1885 (1.0%) | 0.46 | 2/436 (0.5%) | 0.21 |
| Yes | 56/4570 (1.2%) | 10/866 (1.1%) | |||
| VDRL (syphilis test) | Negative | 79/6732 (1.2%) | 0.34 | 16/1282 (1.2%) | 0.71 |
| Positive | 1/34 (2.9%) | 0/11 (0) | |||
| Other risk exposures | |||||
| Substance use | No | 93/7459 (1.2%) | 0.75 | 15/1396 (1.1%) | 0.26 |
| Yes | 2/201 (1.0%) | 1/31 (3.2%) | |||
| Contact with cattle | No | 87/6907 (1.3%) | 0.99 | 14/1318 (1.1%) | 0.13 |
| Yes | 5/399 (1.2%) | 2/64 (3.1%) | |||
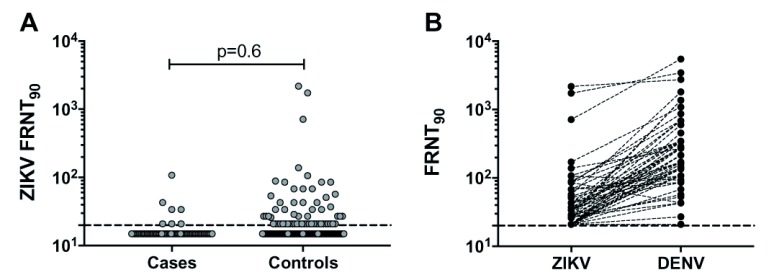
Of the 94 cases with samples available, 71 had sufficient cord plasma material for ZIKV FRNT 90 assay. For the FRNT 90 assay, each of the 71 cases were matched to at least 10 controls by year of birth (n=755). The overall ZIKV FRNT 90 antibody prevalence in the study population was 8.2% (95% CI 6.54, 10.32), but this showed no association with microcephaly ( Table 2 and Figure 3).
Figure 3. DENV and ZIKV neutralising antibody titres in cord plasma.
FRNT 90 antibody titres measured against ZIKV MR766 strain in cord plasma from cases and controls are shown in ( A), including p value from statistical comparison using the Mann-Whitney U test. For ZIKV FRNT 90 seropositive samples (n=68) the corresponding FRNT 90 antibody titres against a local DENV-2 isolate are shown in ( B). The dashed line represents the assay limit of detection.
To characterise the ZIKV FRNT 90 response further we measured FRNT 90 antibody levels of the ZIKV FRNT 90 seropositive cord plasma (7 cases, 61 controls) against a local DENV-2 isolate and compared these to the corresponding ZIKV FRNT 90 titres. All ZIKV seropositive samples had antibody against DENV, with DENV FRNT 90 titres for most samples (43 of the 68 ZIKV seropositive samples) being at least fourfold higher than the corresponding ZIKV FRNT 90 titres ( Figure 3). No sample had a ZIKV FRNT 90 titre that was at least fourfold higher than that for DENV suggesting that the ZIKV response observed in our study population was largely due to cross-reactive responses to DENV or other related flaviviruses. Raw data used in these analyses are available as Underlying data 19.
Discussion
This study set out to estimate the prevalence of microcephaly in coastal Kenya in a cohort of babies born in a rural public county hospital using data from two prospective studies that estimated gestational age clinically and using ultrasound, respectively. We then sought to identify risk factors for microcephaly using a nested case-control design 10. We found a prevalence of 1 to 2%, which given the 0.1% expected prevalence (<-3 SD in the reference populations) suggests a high, previously unrecognised, burden of microcephaly in this region. Using logistic regression we found that newborns with microcephaly were more likely to be born SGA. Finally, we did not detect any ZIKV or flavivirus RNA among cases or controls; nor differences in anti-ZIKV antibody responses between cases and controls.
A similarly high prevalence of microcephaly has been observed in Nigeria where approximately 1.9% of infants had a HC Z score <-3 SD 20. In comparison, the average pre-ZIKV epidemic prevalence of microcephaly in 55 hospitals in the Americas was estimated at 0.04% (95% CI 0.041, 0.049) 21, while the prevalence among 24 European Surveillance of Congenital Anomalies registries ranged between 0.004% and 0.04% 22. During the ZIKV epidemic the overall microcephaly prevalence (HC Z score <-3 SD) in Pernambuco, one of the most severely affected states in Brazil, was estimated at 0.15% (95% CI 0.12, 0.17) 23, almost ten times lower than that observed in our dataset.
Definitions of microcephaly are complex and confounded by prematurity. Could we have overestimated the prevalence? We showed that use of the LMP led to an overestimate of gestational age compared with the gold-standard method of ultrasound scans in early pregnancy, which may have led to an overestimate in the prevalence of microcephaly. Furthermore, errors in measuring head circumference could lead to misclassification. However, when we restricted analysis to the scanned cohort, in which gestational ages were calculated by high quality ultrasound scans in early pregnancy, and where two anthropometrists confirmed head circumferences, the prevalence of microcephaly remained high. Furthermore, even after restricting analysis in the LMP cohort to newborns with birth weight ≥2500 g, thus excluding all low birth weight (and hence most preterm births), the prevalence of microcephaly remained high.
We undertook detailed serological testing in cases and controls and did not find any ZIKV or flavivirus RNA. In addition, we showed no differences in anti-ZIKV FRNT 90 or IgM antibody response between cases and controls. Notably, all the ZIKV FRNT 90 seropositive samples were also seropositive for DENV. Comparison of FRNT 90 titres is conventionally used to infer specific flavivirus exposure 24; if the detected responses were due to recent ZIKV exposure we would expect ZIKV FRNT 90 titres to be at least fourfold higher than the DENV FRNT 90 titres in the corresponding sample(s) 24. However, no sample had a ZIKV FRNT 90 titre that was fourfold higher than the corresponding DENV FRNT 90 titre. In fact, for more than half (63%) of the seropositive samples DENV FRNT 90 titres were at least fourfold higher than ZIKV FRNT 90 titres. Furthermore, the lack of seasonality in the risk of microcephaly, lack of trend by calendar year, and absence of any difference by urban/rural residence suggest it is unlikely that a vector borne or respiratory infection, including ZIKV, is the cause of microcephaly in our setting. The only strongly significant risk factor for microcephaly in our population was SGA; this sub-group accounted for 45% of all cases.
It is possible that we missed the viraemia in cord blood as a result of infection occurring earlier in pregnancy, as is common for ZIKV-associated microcephaly 1, 25. Sample collection in this study was only done at the time of delivery 10. However, if ZIKV was a significant cause of microcephaly we would expect an increase in IgM levels to ZIKV in cord blood, as reported elsewhere among children with microcephaly secondary to ZIKV 26– 29. Furthermore, although molecular evidence of infection is not common among newborns with ZIKV-induced microcephaly, we would expect at least some to have prolonged ZIKV viraemia if this were a common cause. The complete absence of any molecular evidence of ZIKV in cases and controls, including in newborns with measurable cord blood anti-ZIKV FRNT 90 and IgM antibody responses, leads us to believe that the anti-ZIKV response detected in our study is cross-reactive to other flaviviruses. This hypothesis is supported by the high seroprevalence of DENV antibodies measured FRNT 90 in this study and by other methods in previous studies in coastal Kenya 7– 9, 30. Other potential infectious and non-infectious causes 31, 32, including genetic, nutritional and environmental factors, also warrant further investigation and may underlie the associations observed between microcephaly and SGA newborns (as observed by others 29).
In addition to using LMP in a subgroup to define gestational age, this study had other limitations. The study population only included births at KCH, and hence will have missed births occurring at home or in other local health facilities. However, we have important data from our uniquely detailed demographic surveillance system which indicates that approximately 40% of all births in the hospital catchment area occur at KCH.
This study has allowed the first estimation of the risk of congenital microcephaly in coastal Kenya. A 1-2% prevalence of microcephaly may impose a public health burden depending on the clinical outcomes associated. Future prospective studies to characterise and determine post-discharge mortality, neurocognitive outcomes and aetiology of microcephaly in the region are a priority.
Data availability
Underlying data
Harvard Dataverse: Replication Data for: Congenital Microcephaly Unrelated to Flavivirus Exposure in Coastal Kenya. https://doi.org/10.7910/DVN/4EB9PG 19.
This project contains the following underlying data:
microcephaly_dataverse_v1 (dataset containing demographic information, anthropometric measures and results of lab assays for participants included in the study).
GWarimwe_Microcephaly_Codebook (contains variable description and value labels).
Extended data
Harvard Dataverse: Replication Data for: Congenital Microcephaly Unrelated to Flavivirus Exposure in Coastal Kenya. https://doi.org/10.7910/DVN/4EB9PG 19.
This project contains the following extended data:
GWarimwe_Microcephaly_missing_data_summary (summary of missing data for analysed variables).
GWarimwe_Microcephaly_readme (readme file).
microcephaly_dataverse_final (STATA analysis code used for data analysis presented in this article).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Acknowledgements
We are grateful to Grieven Otieno and Mark Otiende for help with data management and sample access, and to the KIPMAT team for participant recruitment and evaluation. We are grateful to the INTERBIO-21 st Study coordination team for training, assessment and monitoring of ultrasound scanning and newborn anthropometry.
Funding Statement
This work was supported through an award from the Wellcome Trust (grant no. 203077_Z_16_Z), the INTERGROWTH-21st grant no. 49038 from the Bill & Melinda Gates Foundation, and an award from the Medical Research Council, Wellcome Trust, and Newton Fund (grant no. MC_PC_15092) that is part of the EDCTP2 programme supported by the European Union. This manuscript was submitted for publication with permission from the Director of the Kenya Medical Research Institute.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved, 1 approved with reservations]
References
- 1. Petersen LR, Jamieson DJ, Powers AM, et al. : Zika Virus. N Engl J Med. 2016;374(16):1552–63. 10.1056/NEJMra1602113 [DOI] [PubMed] [Google Scholar]
- 2. Dick GW: Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46(5):521–34. 10.1016/0035-9203(52)90043-6 [DOI] [PubMed] [Google Scholar]
- 3. Geser A, Henderson BE, Christensen S: A multipurpose serological survey in Kenya. 2. Results of arbovirus serological tests. Bull World Health Organ. 1970;43(4):539–52. [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson BK, Musoke S, Ocheng D, et al. : Dengue-2 virus in Kenya. Lancet. 1982;2(8291):208–9. 10.1016/s0140-6736(82)91047-9 [DOI] [PubMed] [Google Scholar]
- 5. Vu DM, Mutai N, Heath CJ, et al. : Unrecognized Dengue Virus Infections in Children, Western Kenya, 2014-2015. Emerg Infect Dis. 2017;23(11):1915–7. 10.3201/eid2311.170807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellis BR, Barrett AD: The enigma of yellow fever in East Africa. Rev Med Virol. 2008;18(5):331–46. 10.1002/rmv.584 [DOI] [PubMed] [Google Scholar]
- 7. Mease LE, Coldren RL, Musila LA, et al. : Seroprevalence and distribution of arboviral infections among rural Kenyan adults: a cross-sectional study. Virol J. 2011;8:371. 10.1186/1743-422X-8-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ochieng C, Ahenda P, Vittor AY, et al. : Seroprevalence of Infections with Dengue, Rift Valley Fever and Chikungunya Viruses in Kenya, 2007. PLoS One. 2015;10(7):e0132645. 10.1371/journal.pone.0132645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sutherland LJ, Cash AA, Huang YJ, et al. : Serologic evidence of arboviral infections among humans in Kenya. Am J Trop Med Hyg. 2011;85(1):158–61. 10.4269/ajtmh.2011.10-0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seale AC, Barsosio HC, Koech AC, et al. : Embedding surveillance into clinical care to detect serious adverse events in pregnancy. Vaccine. 2015;33(47):6466–8. 10.1016/j.vaccine.2015.07.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kennedy SH, Victora CG, Craik R, et al. : Deep clinical and biological phenotyping of the preterm birth and small for gestational age syndromes: The INTERBIO-21 st Newborn Case-Control Study protocol [version 2; peer review: 2 approved]. Gates Open Res. 2019;2:49. 10.12688/gatesopenres.12869.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patel P, Landt O, Kaiser M, et al. : Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013;10:58. 10.1186/1743-422X-10-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corman VM, Rasche A, Baronti C, et al. : Assay optimization for molecular detection of Zika virus. Bull World Health Organ. 2016;94(12):880–92. 10.2471/BLT.16.175950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. World Health Organization: Screening, assessment and management of neonates and infants with complications associated with Zika virus exposure in utero.WHO/ZIKV/MOC/163/Rev3.2016. Reference Source [Google Scholar]
- 15. Villar J, Giuliani F, Fenton TR, et al. : INTERGROWTH-21st very preterm size at birth reference charts. Lancet. 2016;387(10021):844–5. 10.1016/S0140-6736(16)00384-6 [DOI] [PubMed] [Google Scholar]
- 16. Villar J, Cheikh Ismail L, Victora CG, et al. : International standards for newborn weight, length, and head circumference by gestational age and sex: the Newborn Cross-Sectional Study of the INTERGROWTH-21 st Project. Lancet. 2014;384(9946):857–68. 10.1016/S0140-6736(14)60932-6 [DOI] [PubMed] [Google Scholar]
- 17. Ngoi CN, Price MA, Fields B, et al. : Dengue and Chikungunya Virus Infections among Young Febrile Adults Evaluated for Acute HIV-1 Infection in Coastal Kenya. PLoS One. 2016;11(12):e0167508. 10.1371/journal.pone.0167508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kamau E, Agoti CN, Ngoi JM, et al. : Complete Genome Sequences of Dengue Virus Type 2 Strains from Kilifi, Kenya. Microbiol Resour Announc. 2019;8(4):pii: e01566–18. 10.1128/MRA.01566-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barsosio HC, Gitonga JN, Karanja HK: Replication Data for: Congenital Microcephaly Unrelated to Flavivirus Exposure in Coastal Kenya. Harvard Dataverse, V2.2019. 10.7910/DVN/4EB9PG [DOI] [PMC free article] [PubMed]
- 20. Olusanya BO: Full-term newborns with congenital microcephaly and macrocephaly in Southwest Nigeria. Int Health. 2012;4(2):128–34. 10.1016/j.inhe.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 21. Orioli IM, Dolk H, Lopez-Camelo JS, et al. : Prevalence and clinical profile of microcephaly in South America pre-Zika, 2005-14: prevalence and case-control study. BMJ. 2017;359:j5018. 10.1136/bmj.j5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morris JK, Rankin J, Garne E, et al. : Prevalence of microcephaly in Europe: population based study. BMJ. 2016;354:i4721. 10.1136/bmj.i4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kleber de Oliveira W, Cortez-Escalante J, De Oliveira WT, et al. : Increase in Reported Prevalence of Microcephaly in Infants Born to Women Living in Areas with Confirmed Zika Virus Transmission During the First Trimester of Pregnancy - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(9):242–7. 10.15585/mmwr.mm6509e2 [DOI] [PubMed] [Google Scholar]
- 24. World Health Organization: Laboratory testing for Zika virus infection.WHO/ZIKV/LAB/161.2016. Reference Source [Google Scholar]
- 25. Cauchemez S, Besnard M, Bompard P, et al. : Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet. 2016;387(10033):2125–32. 10.1016/S0140-6736(16)00651-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cordeiro MT, Pena LJ, Brito CA, et al. : Positive IgM for Zika virus in the cerebrospinal fluid of 30 neonates with microcephaly in Brazil. Lancet. 2016;387(10030):1811–2. 10.1016/S0140-6736(16)30253-7 [DOI] [PubMed] [Google Scholar]
- 27. Cordeiro MT, Brito CA, Pena LJ, et al. : Results of a Zika Virus (ZIKV) Immunoglobulin M-Specific Diagnostic Assay Are Highly Correlated With Detection of Neutralizing Anti-ZIKV Antibodies in Neonates With Congenital Disease. J Infect Dis. 2016;214(12):1897–904. 10.1093/infdis/jiw477 [DOI] [PubMed] [Google Scholar]
- 28. Lanciotti RS, Kosoy OL, Laven JJ, et al. : Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14(8):1232–9. 10.3201/eid1408.080287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Araújo TVB, Ximenes RAA, Miranda-Filho DB, et al. : Association between microcephaly, Zika virus infection, and other risk factors in Brazil: final report of a case-control study. Lancet Infect Dis. 2018;18(3):328–36. 10.1016/S1473-3099(17)30727-2 [DOI] [PubMed] [Google Scholar]
- 30. Geser A, Christensen S, Thorup I: A multipurpose serological survey in Kenya. 1. Survey methods and progress of field work. Bull World Health Organ. 1970;43(4):521–37. [PMC free article] [PubMed] [Google Scholar]
- 31. Devakumar D, Bamford A, Ferreira MU, et al. : Infectious causes of microcephaly: epidemiology, pathogenesis, diagnosis, and management. Lancet Infect Dis. 2018;18(1):e1–e13. 10.1016/S1473-3099(17)30398-5 [DOI] [PubMed] [Google Scholar]
- 32. Nawathe A, Doherty J, Pandya P: Fetal microcephaly. BMJ. 2018;361:k2232. 10.1136/bmj.k2232 [DOI] [PubMed] [Google Scholar]