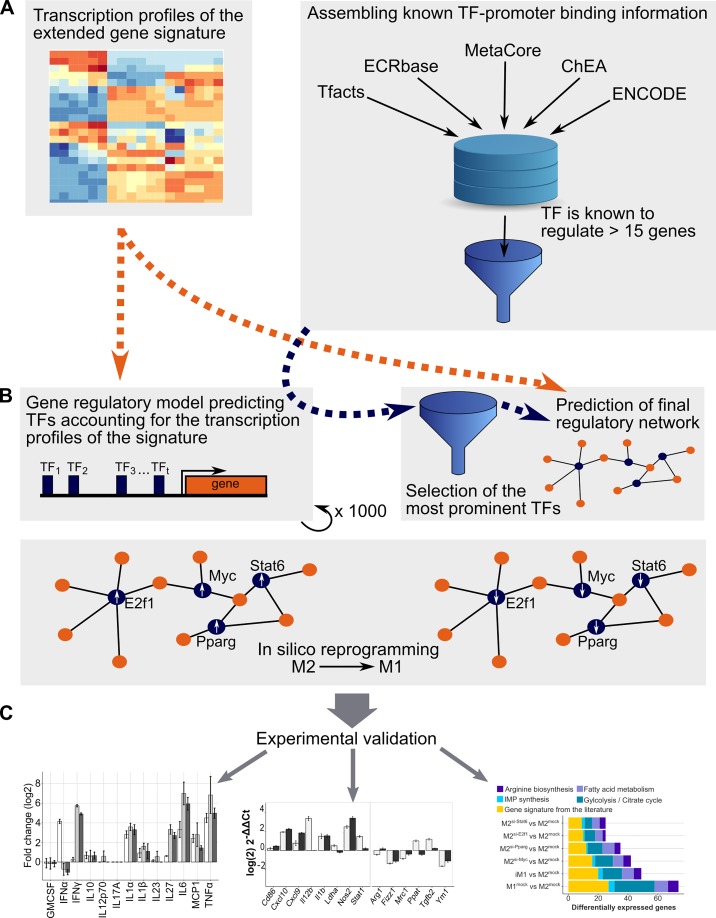
Fig 3. The analysis pipeline to obtain the gene regulatory model.
(A) Gene expression profiles and evidence-based interactions for all transcription factors were collected. (B) After a pre-filtering step, the activity of each transcription factor was determined, and regulatory network models were generated. The transcription factors that were most often selected by the models across all target genes were incorporated into the extended gene signature, a parsimony model constructed, and in silico reprogramming was performed to convert the activities of these transcription factors into M1-like activities. (C) The predicted transcription factors were experimentally validated by investigating the cytokine secretion and transcription profiles of the reprogrammed M2-like macrophages after knocking down the expression of the predicted transcription factors.