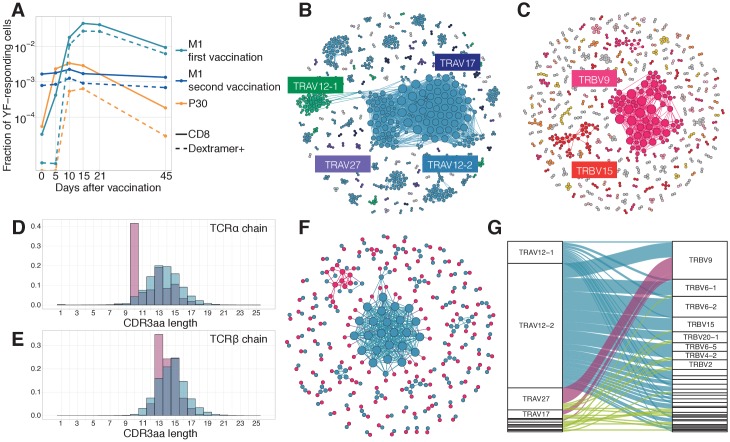
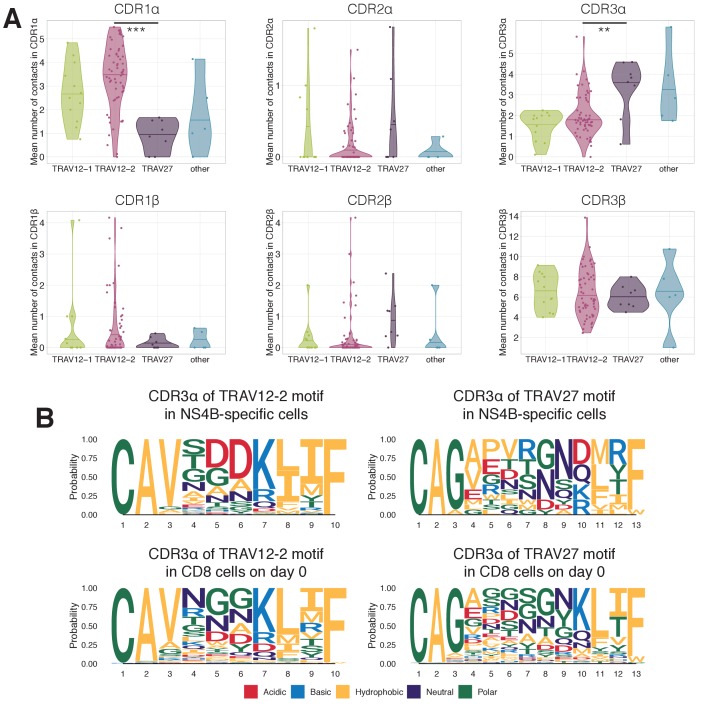
Figure 4. Response to the immunodominant yellow fever epitope NS4B214-222.
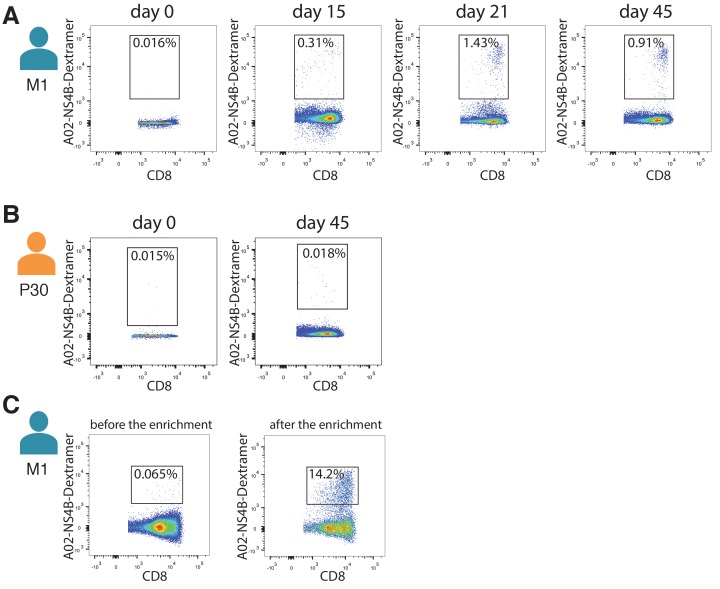
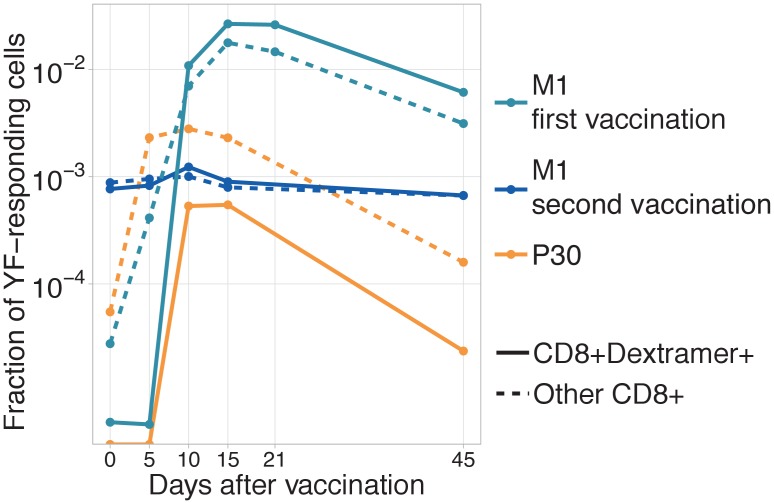
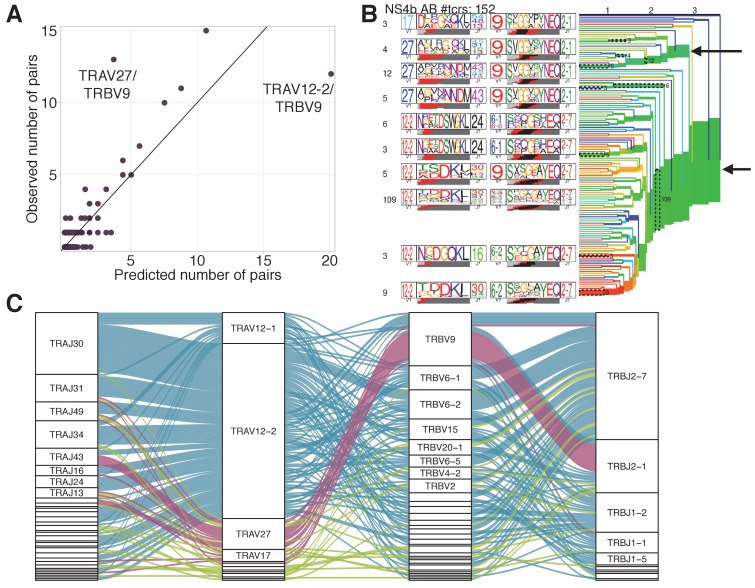
(A) Fraction of all T-cells corresponding to CD8+ YF-responding TCRβ clonotypes (solid lines) and CD8+NS4B-specific clonotypes (dashed lines) as a function of time post-vaccination (x-axis). Sequence similarity networks for TCR alpha (B) and beta (C) of NS4B-positive cells. Each vertex is a TCR amino acid sequence, connected with an edge if they differ by fewer than two mismatches. The size of the vertex indicates its degree. Vertices of zero degree are not shown. Color and text boxes indicate V-segments that are significantly enriched (exact Fisher test, Benjamini Hochberg adjusted ) in usage in epitope-specific cells compared to the bulk repertoire. NS4B-specific TCR alpha (D) and TCR beta (E) chains (red histograms) have biases in CDR3 length in comparison to bulk TCR repertoire of CD8+ cells (overlayed blue histograms). (F) Network of single-cell paired TCR alpha (blue) and TCR beta (red) of NS4B-specific TCRs. Vertices of the same color are connected if there are less than two mismatches in TCR chain amino acid sequence. An edge between vertices of different color represents the pairing of alpha and beta. The biggest alpha cluster (blue in the center) corresponds to the TRAV12-2 cluster on B, and it pairs with many dissimilar beta chains. The biggest beta cluster (top left in red) corresponds to the TRBV9 cluster of C. (G) Pairing of V-segments of TCR alpha (left) to V-segments of TCR beta (right) in scTCRseq of NS4B-specific T-cells. The height of each box is proportional to the number of unique clones with this V-segment. The width of ribbons is proportional to the frequency of TRAV-TRBV combination. NS4B-specific TCRs have two main binding modes, defined by TRAV12 segment family paired to almost any TRBV (blue) and by TRAV27 segment paired preferentially with TRBV9 (pink).