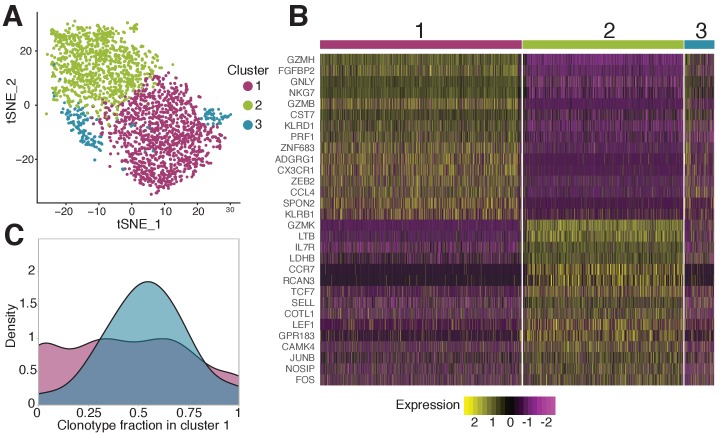
Figure 5. Phenotypic diversity of NS4B-specific cells 18 months after yellow fever immunization.
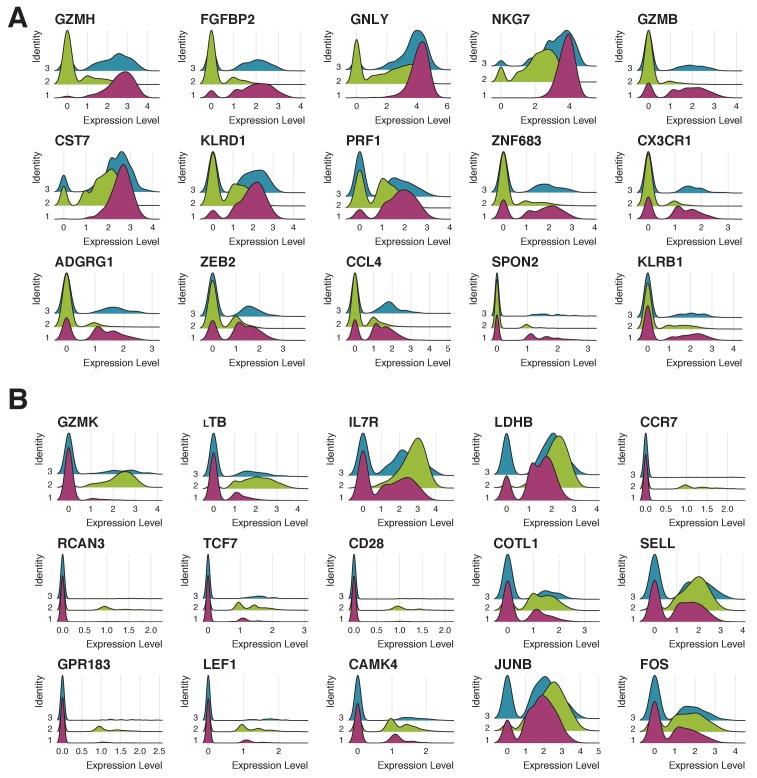
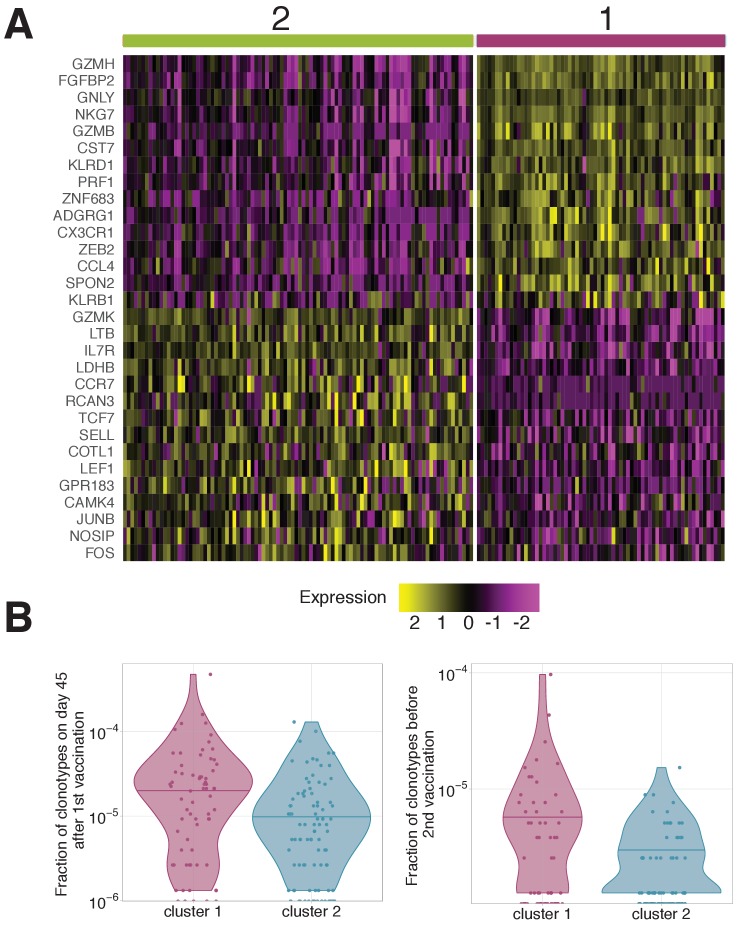
(A) 2D t-SNE visualization of unsupervised clustering (Seurat analysis) of RNAseq data based on 2000 most variable genes shows three distinct clusters of NS4B-specific cells. (B) The heatmap of top 15 significantly enriched genes of single cells in clusters 1 and 2 defined by the MAST algorithm. The panel above the heatmap identifies the cluster identity of the cells. (C) Gaussian kernel density estimate for the relative fraction of cells belonging to cluster one for each clonotype. Blue distribution shows the theoretical prediction under the null hypothesis: clonotype labels were shuffled between cells (1000 permutations). The observed distribution is flatter than the theoretical one, indicating the presence of clonotypes with either a minority or a majority of cells belonging to cluster 1 ( -test with MC-estimated p-value=0.0005).