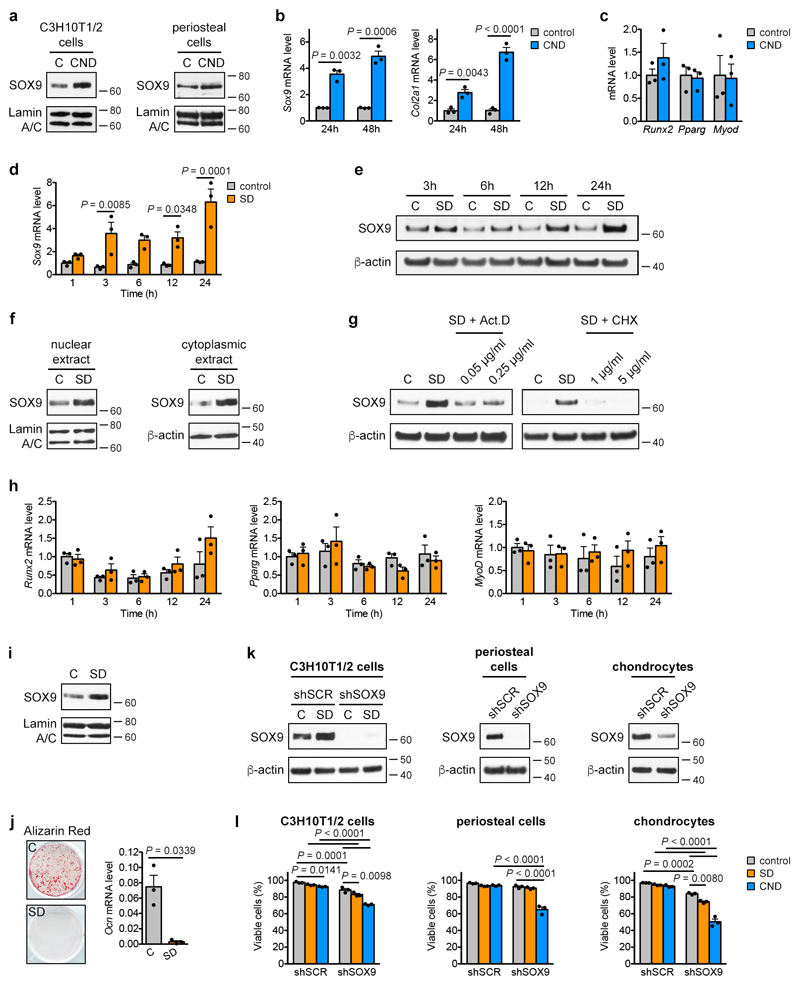
Extended Data Figure 4. Skeletal progenitors resist nutritional stress via induction of SOX9.
(a) Immunoblot detection of nuclear SOX9 in C3H10T1/2 cells and periosteal cells exposed for 24 hours to control or CND medium, with Lamin A/C as loading control (n=2 independent experiments). (b) mRNA levels of Sox9 and Col2a1 in periosteal cells exposed for the indicated times to control or CND medium (relative to control, n=3 biologically independent samples). (c) mRNA levels of runt-related transcription factor 2 (Runx2; osteogenic lineage), peroxisome proliferator-activated receptor γ (Pparg; adipogenic lineage) and MyoD (myogenic lineage) in periosteal cells exposed for 48 hours to control or CND medium (relative to control, n=3 biologically independent samples). (d) mRNA levels of Sox9 in C3H10T1/2 cells exposed for the indicated times to control or SD medium (relative to control, n=3 independent experiments). (e) Immunoblot detection of total SOX9 in C3H10T1/2 cells exposed for different durations to control or SD medium, with β-actin as loading control (n=2 independent experiments). (f) Immunoblot detection of nuclear and cytoplasmic SOX9 in C3H10T1/2 cells exposed for 6 hours to control or SD medium, with Lamin A/C or β-actin as loading control (n=2 independent experiments). (g) Immunoblot detection of SOX9 in total cell protein extracts of C3H10T1/2 cells exposed for 6 hours to control medium, SD medium or SD medium supplemented with different concentrations of the transcription inhibitor Actinomycin D (Act. D) or the translation inhibitor cycloheximide (CHX). Detection of β-actin was used as loading control (n=2 independent experiments). (h) mRNA levels of Runx2, Pparg and MyoD in C3H10T1/2 cells exposed for the indicated times to control or SD medium (relative to control, n=3 independent experiments). (i) Immunoblot detection of nuclear SOX9 in periosteal cells exposed for 24 hours to control or SD medium with Lamin A/C as loading control (n=3 biologically independent samples). (j) Osteogenic differentiation of periosteal cells in control or SD medium, assessed by visualization of mineral deposits (Alizarin Red staining) and quantification of Ocn mRNA levels (relative to Actin, n=3 biologically independent samples). (k) Immunoblot detection of SOX9 in total cell protein extracts of C3H10T1/2 cells (in control or SD medium), periosteal cells and growth plate-derived chondrocytes transduced with shSOX9 or shSCR, with β-actin as loading control. A longer exposure time was used for SOX9 detection in C3H10T1/2 cells and periosteal cells compared to chondrocytes in order to visualize any remaining protein in the shSOX9 conditions (n=2 independent experiments for C3H10T1/2 cells, n=3 biologically independent samples for periosteal cells, growth plate-derived chondrocytes). (l) Quantification of cell viability of C3H10T1/2 cells, periosteal cells and growth plate-derived chondrocytes transduced with shSOX9 or shSCR, after 72 hours of exposure to control, SD or CND medium (n=3 independent experiments for C3H10T1/2 cells, n=3 biologically independent samples for periosteal cells, growth plate-derived chondrocytes). Mean ± s.e.m. Two-way ANOVA with Bonferroni post-hoc test (b,d,h,l), two-tailed Student’s t-test (c,j). For gel source data, see Supplementary Figure 1.