Fig. 4.
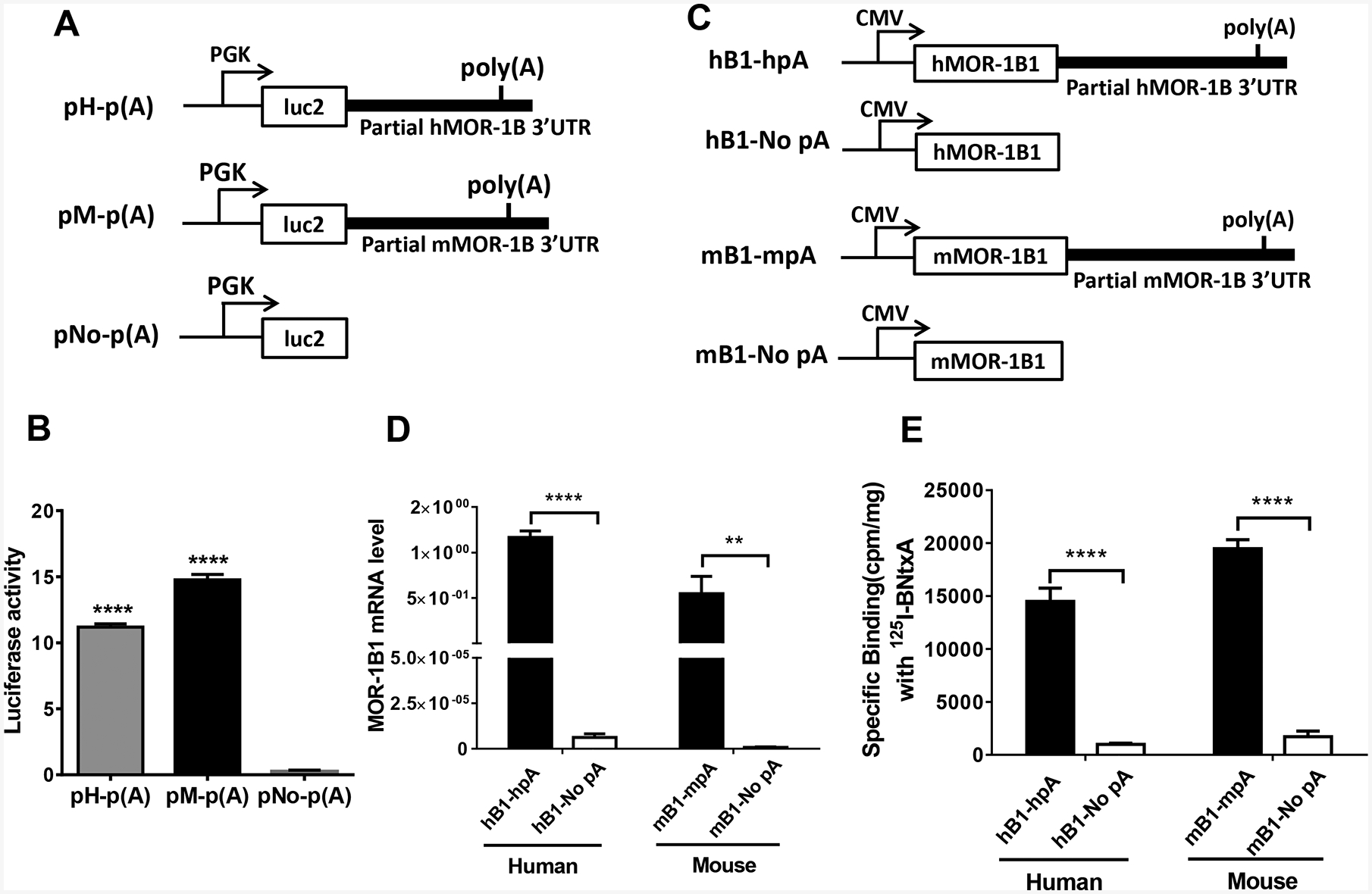
Role of the 3’-UTR of hMOR-1B and mMOR-1B in luciferase activity and MOR-1B1 expression. A). Schematic of the constructs for luciferase assay. The partial 3’-UTR of hMOR-1B and mMOR-1B containing poly(A), cleavage site and U/G-rich region was subcloned downstream of the firefly luciferase (luc2) coding region in pmir vector, as pH-p(A) and pM-p(A), respectively, as described in Materials and Methods. PGK promoter is indicated by arrows.
B). Luciferase activity. The lysates from transfected HEK293T cells with the indicated constructs (n = 4) were used for analyzing luciferase activity by using Dual-Glo Luciferase Assay, as described in Materials and Methods. Luciferase activity was calculated by normalizing with Renilla luciferase activity. Statistically significant differences were calculated by one-way ANOVA with Bonferroni’s post hoc analysis. ****: p < 0.0001 compared with pNo-p(A).
C). Schematic of the MOR-1B1 constructs. The partial 3’-UTR of hMOR-1Bs and mMOR-1Bs containing poly(A), cleavage site and U/G-rich region was subcloned downstream of hMOR-1B1 and mMOR-1B1 coding region in pcDNA3 vector, as hB1-hpA and as mB1-mpA, respectively. hB1-No pA and mB1-No pA were used as controls. CMV promoter is shown by arrows.
D). Expression of MOR-1B1 mRNAs. Total RNAs from transfected HEK293 cells with the indicated constructs (n = 4) were used in RT-qPCR to determine the expression of MOR-1B1 mRNA, as described in Materials and Methods. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001. **: p < 0.01.
E). Opioid receptor binding. Membrane proteins from transfected HEK293 cells with the indicated constructs (n = 4) were used in 125I-IBNtxA binding assay, as described in Materials and Methods. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ****: p < 0.0001.
