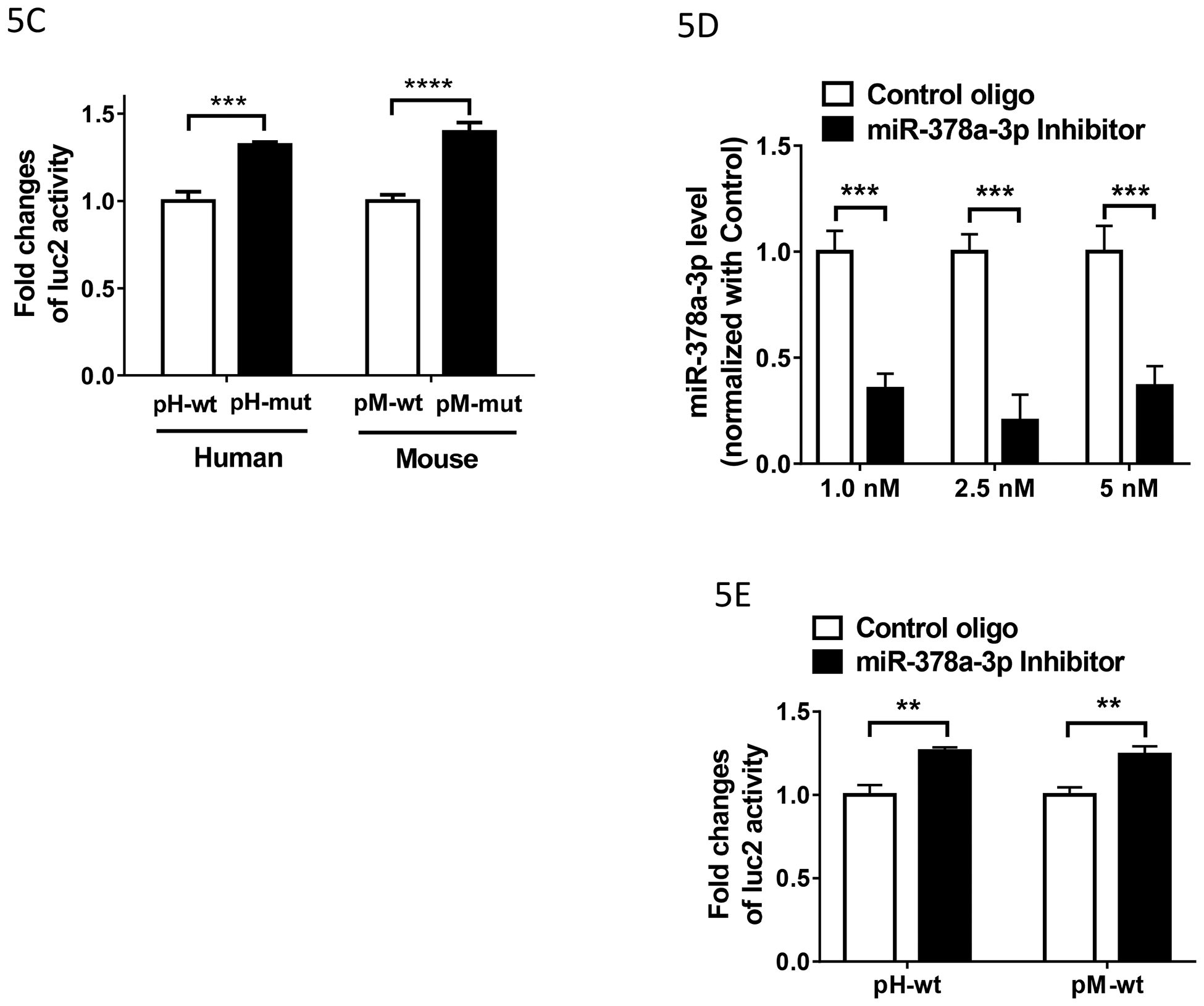
Fig. 5.
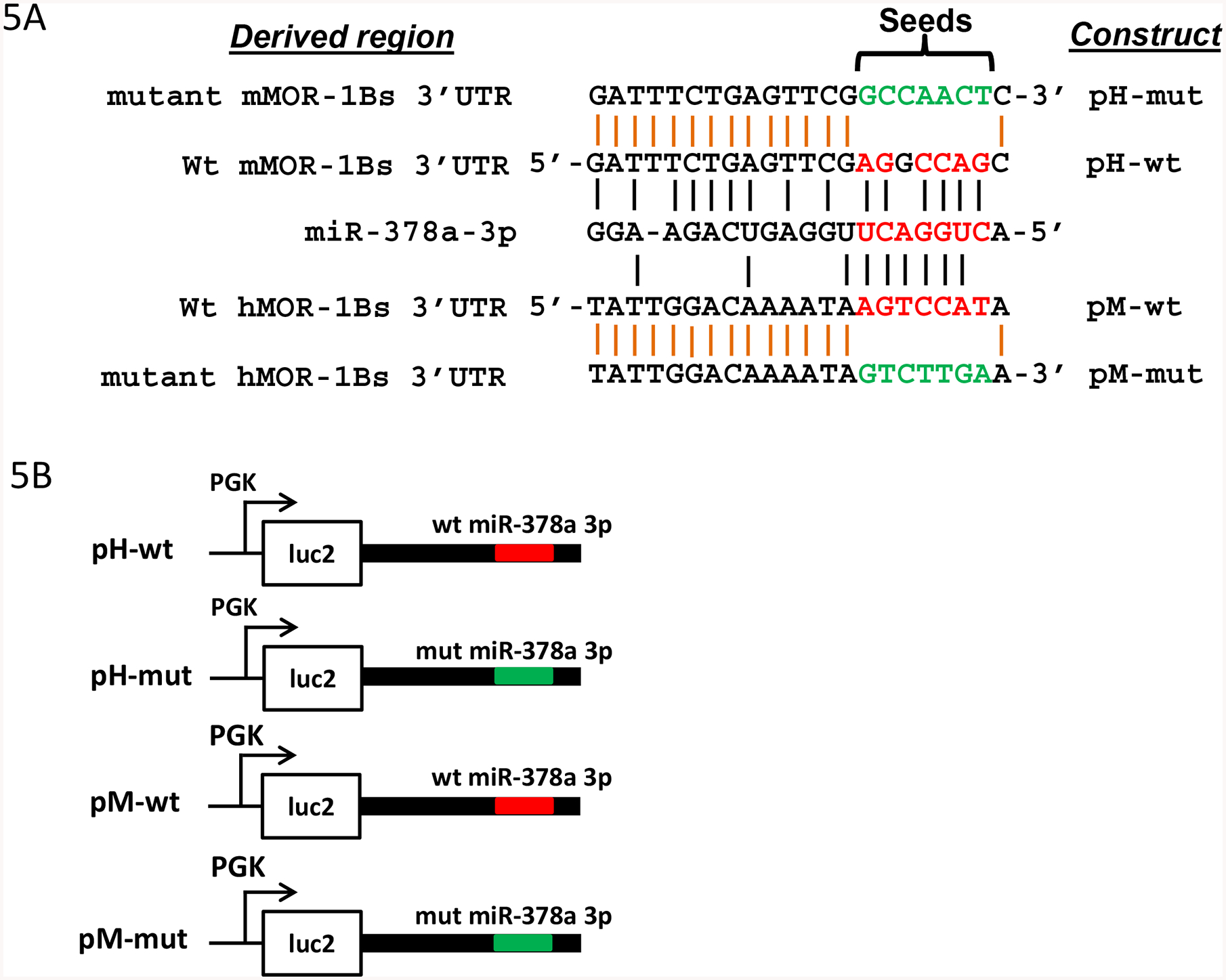
Regulation of luciferase activity by miR-378a-3p through a conserved miR-378a-3p binding site in MOR-1Bs 3’-UTRs. A). Alignment of miR-378a-3p sequences with 3’-UTR of hMOR-1Bs and mMOR-1Bs and mutagenized sequences of the miR-378a-3p sites. Red letter: the miR-378a-3p seeds and aligned 3’-UTR sequences. Green letter: mutagenized sequence. Black vertical lines show the complementary alignment and orange vertical lines indicate identical alignment.
B). Constructs in pmir vector. The 3’-UTRs of hMOR-1Bs and mMOR-1Bs containing wild-type (wt) or mutated (mut) miR-378a-3p sequences in A were subcloned into pmir as pH-wt, pM-wt, pH-mut, and pM-mut, respectively, as described in Materials and Methods. The wt and mut miR-378a-3p sequences are indicated by red and green lines, respectively.
C). Effect of the miR-378a-3p mutation on the luciferase (luc2) activity in HEK293Tcells. Luciferase activity was measured using the lysate from HEK293 cells transfected with indicated constructs (n = 4), as described in Materials and Methods. Fold change of luc2 activity was calculated by normalizing the values of the mutant (mut) constructs with those of the wild-type (wt) constructs. Two ANOVA with Bonferroni’s post hoc analysis was used. ***: p < 0.001; ****: p < 0.0001.
D). Effect of miR-378a-3p inhibitor on the expression of miR-378a-3p in HEK293T cells. Total RNAs including small RNAs were isolated using miRNeasy Kit from HEK293 cells transfected with indicated concentration of Control oligo or miR-378a-3p Inhibitor (n = 3–6) and used in RT-qPCR to determine miR-378a-3p level, as described in Materials and Methods. Inhibition level by the LNA inhibitor was calculated by normalizing the levels with those of Control LNA oligo. Two-way ANOVA with Bonferroni’s post hoc analysis was used. ***: p < 0.001.
E). Effect of miR-378a-3p inhibitor on the luciferase (luc2) activity in HEK293T cells. Luciferase activity was measured using the lysate from HEK293 cells transfected with indicated constructs (n = 4). Fold change of luc2 activity was calculated by normalizing the values of the Inhibitor with those of the Control oligo. Two ANOVA with Bonferroni’s post hoc analysis was used. **: p < 0.01.
