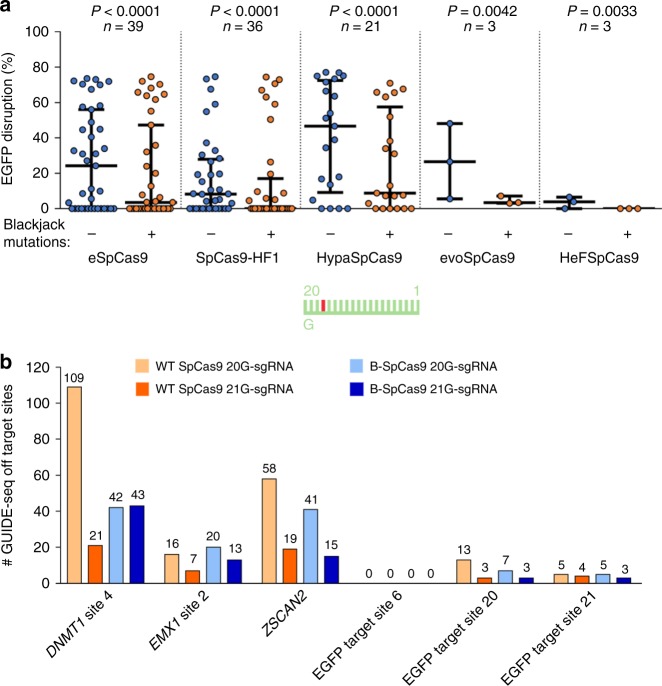
Fig. 3. The Blackjack mutations increase the fidelity of increased fidelity nucleases.
a Blackjack mutations increase the fidelity of their respective parent SpCas9 variants. EGFP-disruption activities with partially mismatching 20G-sgRNAs. Results are shown only for those target sites where both the non-Blackjack parent- and Blackjack-SpCas9 variant exhibit at least 70% on-target activity (with perfectly matching 20G-sgRNAs) compared to WT SpCas9. Only one target (with three mismatched positions) matches this condition in the case of evo- or HeFSpCas9. The median and the interquartile range are shown; data points are plotted as open circles representing the mean of biologically independent triplicates. Spacers are schematically depicted beside the charts as combs: green color teeth indicate matching-, while a red color tooth indicates the presence of a mismatching nucleotide (not necessarily the exact position) within the spacer; numbering of the tooth positions corresponds to the distance of the nucleotide from the PAM; the starting 20th nucleotide of the spacer is indicated by an uppercase letter. Statistical significance was assessed using two-sided Paired-samples Student’s t-test or two-sided Wilcoxon signed ranks test as appropriate; ns not significant. Summary of data distributions and statistical details are reported in Supplementary Data 6. See also Supplementary Fig. 4. b Bar chart of the total number of off-target sites detected by GUIDE-seq for WT and B-SpCas9 variants on six target sites targeted with 20G- or 21G-sgRNAs. See also Supplementary Fig. 5.