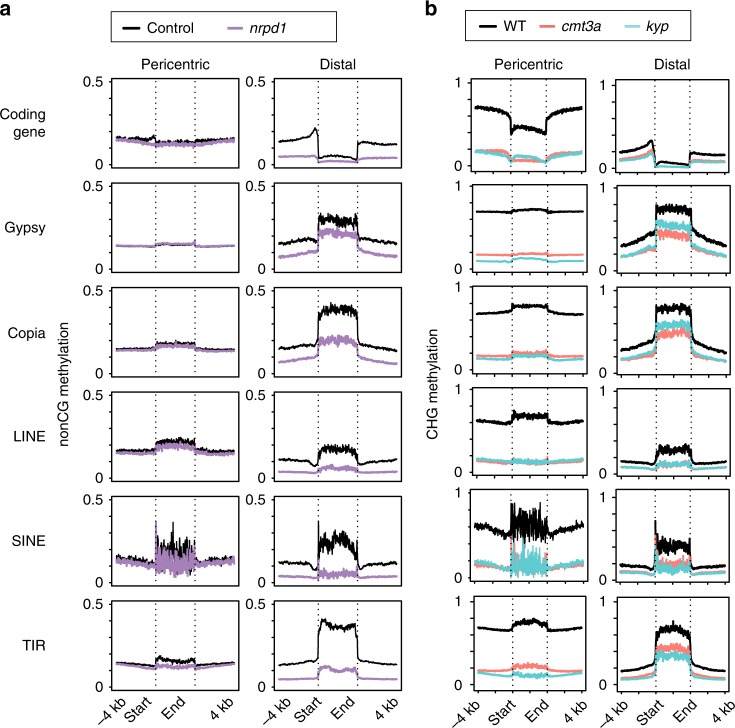
Fig. 2. DNA methylation of TEs in pericentric and distal chromatin.
a Average non-CG methylation over coding genes and different TE families in control and nrpd1. b Average CHG methylation in WT, kyp and cmt3a. Distal and pericentric chromatins are plotted separately. Different genomic elements are aligned at the 5′ end or the 3′ end, and average methylation for all cytosines within each 100 bp interval is plotted. The dotted lines represent the points of alignments of coding-gene transcriptional start site or annotated repeat elements start site and coding-gene transcription termination site or annotated repeat elements end site, respectively. TIR, terminal inverted repeats; LINE, long interspersed nuclear elements; SINE, short interspersed nuclear elements.