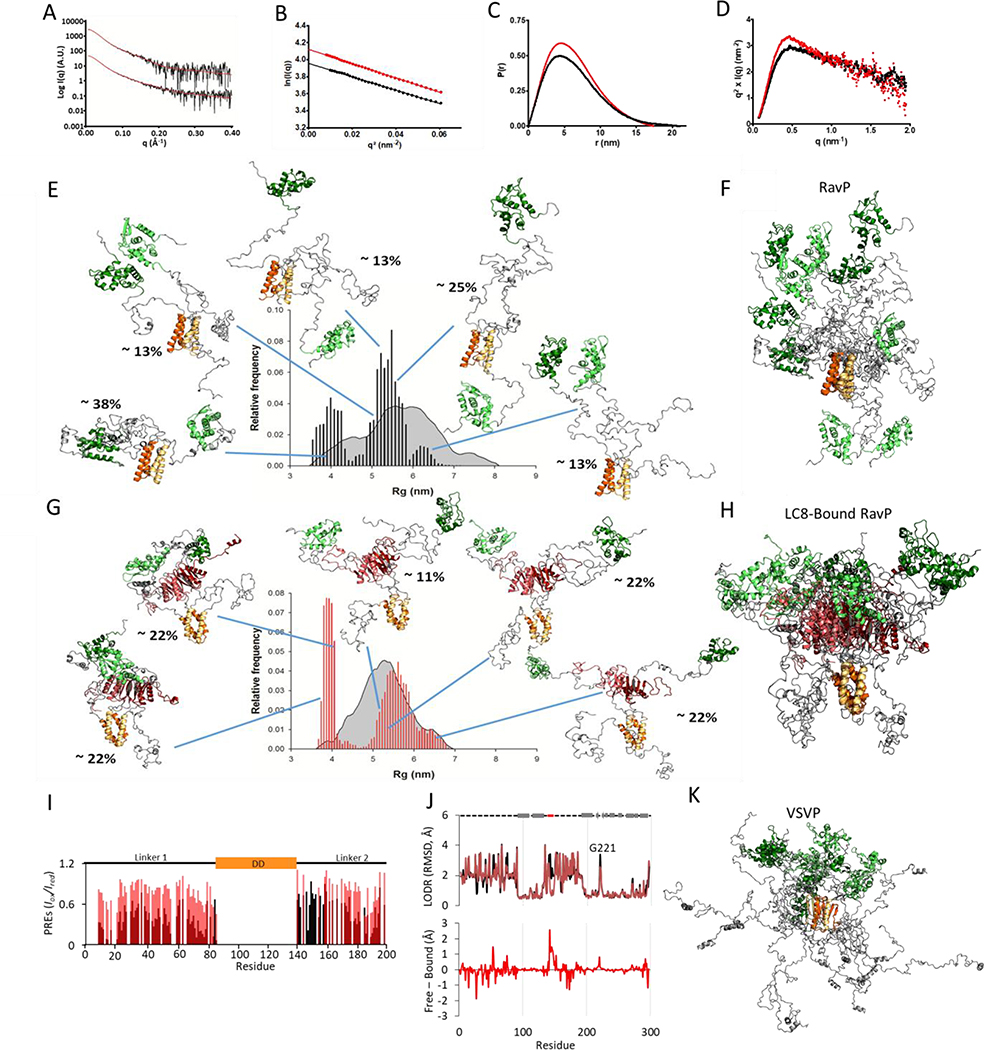
Fig. 4. SAXS data and selected ensembles analysis.
(A) Fitted SAXS profiles for RavP (bottom) and LC8-RavP (top). The experimental curve is shown as a black line and the EOM selected ensemble as a red line. (B) Guinier plot for RavP (in black) and LC8-RavP complex (in red). (C) Pair distribution function for RavP (in black) and LC8-RavP complex (in red). (D) Kratky plot for RavP (in black) and LC8-RavP complex (in red). (E) Radius of gyration distributions for the initial pool (grey) and EOM-selected RavP ensembles (black histogram). RavP models from the EOM are shown as cartoon with their central dimerization domains colored in orange and their C-terminal domains in green. The intrinsically disordered regions are shown in grey. The relative contribution of each model to the EOM calculated SAXS profile is indicated as a percentage. The RavP selected ensemble is overlaid and aligned on the DD in (F). (G) Radius of gyration distributions for the initial pool (grey) and EOM-selected LC8-RavP ensembles (red histogram). Models are shown with LC8 in red, and overlaid in (H). (I) PRE intensity ratios of the paramagnetic (oxidized) to diamagnetic (reduced) of MTSL labelled Rav-C261S free (black) or RavP-C261-LC8 complex (red). Values are only shown for residues 1–196, as the CTD peaks were not visible in this experiment. (J, Top) Ensemblator analysis of models generated via SBM and MDS for RavP (black) and LC8-RavP (red). Models for each sample were compared in one group, with automatic clustering turned off. Residues with a higher LODR in the complex represent regions that are less restricted in motion when LC8 is bound to RavP. Lower LODR regions, on the other hand, are those that are more rigidified within the complex ensemble. A comparison of the differences is shown below. Note the large decrease in variability for residues in the LC8 binding site (residues 140–149). Differences in the CTD, particularly in the loop containing residue 221, are distal from the LC8 site and therefore unexpected. (K) VSVP ensemble overlay of 5 structures, adapted from Leyrat et al., 2012 (23), with conformers aligned using the DD.