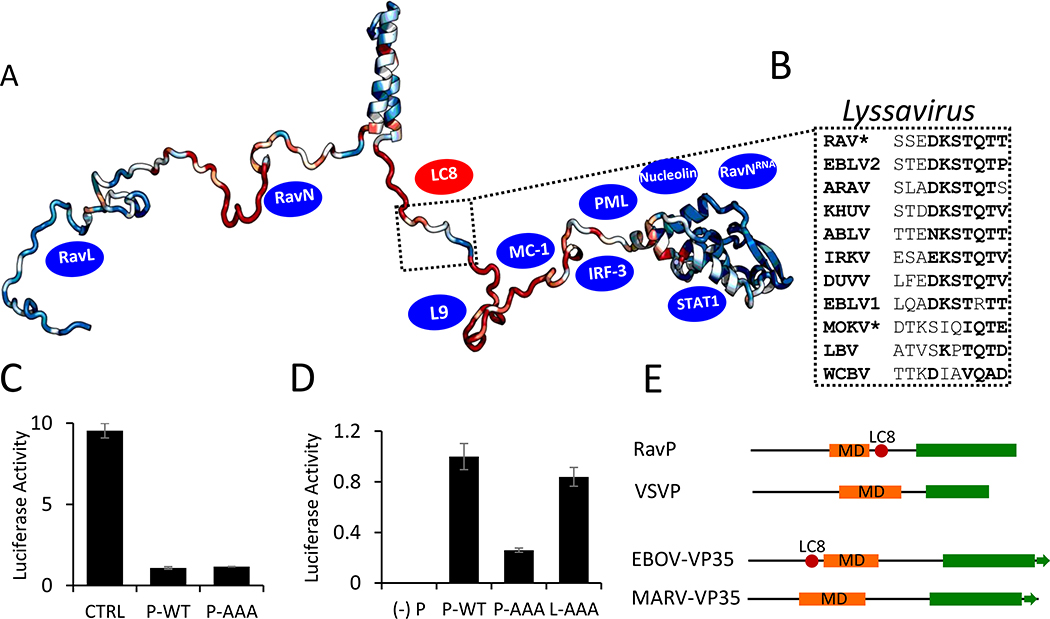
Fig. 5. Phylogenetic and functional analysis of the Rhabdoviridae family and Lyssavirus genus Phosphoprotein.
(A) ConSurf analysis of P proteins within the Lyssavirus genus, with conserved residues shown in blue, and variable residues shown in red, mapped onto a EOM-generated RavP protomer. The LC8 binding sequence is highlighted, with example sequences from viruses within the Lyssavirus genus shown in (B). Residues in bold are amino acids frequently present in LC8 binding partners. *Denotes sequences verified to bind LC8 (1). Blue ovals denote approximate binding sites of known partners (see Discussion). (C) Luciferase reporter assays for IFNα-treated Hek-293T cells (2,000 U/mL, 6 hours). Cells were transfected with luciferase assay components, as well as a plasmid encoding RavP-WT, RavP-AAA, or an empty vector control. (D) Luciferase expression from the RAV minigenome system. BSR-T7 cells were transfected with either RavP constructs shown, as well as RavN, RavL or RavL-AAA, and luciferase reporter assay components. All luciferase values are normalized to the RavP-WT transfection, with error bars shown as the standard deviation from the mean for two independent experiments, run in triplicate. A negative control is shown ((−) P), in which the RavP plasmid was not transfected. (E) Predicted domain architectures for RavP, VSVP, Ebola virus VP35, and Marburg virus VP35. LC8 sites are marked by red ovals.