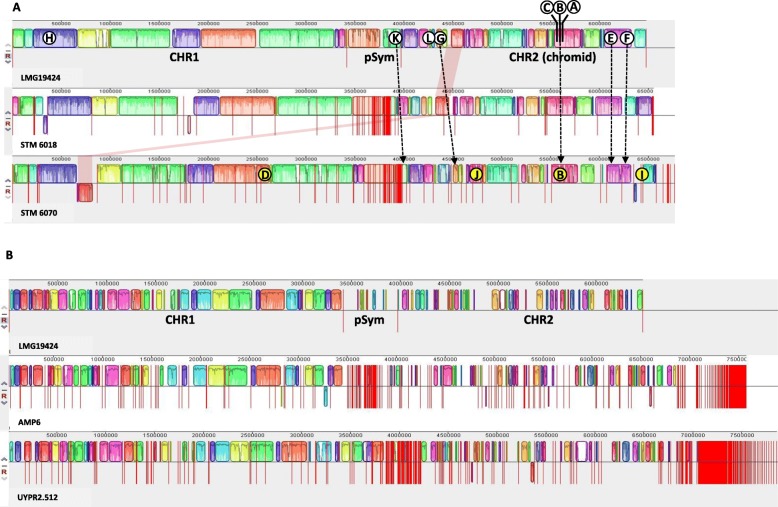
Fig. 1.
Genome alignments using progressive Mauve software [44]. a: scaffolds of the draft genomes of Cupriavidus neocaledonicus STM 6070 (STM 6070) and C. taiwanensis STM 6018 aligned to the replicons of the finished genome of Cupriavidus taiwanensis LMG 19424T (LMG 19424). b: scaffolds of the draft genomes of Cupriavidus sp. strains AMP6 and UYPR2.512 aligned to the replicons of the finished genome of Cupriavidus taiwanensis LMG 19424T (LMG 19424). The blocks in the alignment represent the common local colinear blocks (LCBs) among the compared genomes, and homologous blocks in each genome are shown as identical coloured regions. The vertical red lines represent replicon boundaries for LMG 19424T, whereas they represent contig boundaries for the draft genomes. The shaded red region represents a putative genomic rearrangement between CHR2 and CHR1. Circles with numbers represent the location of heavy metal resistance regions identified in this paper found in LMG 19424T (white circles containing letters) and in STM 6070 (yellow circles containing letters). See Fig. 3 for the heavy metal resistance regions. Dashed arrows show the location of the LMG 19424T heavy metal resistance regions in STM 6070