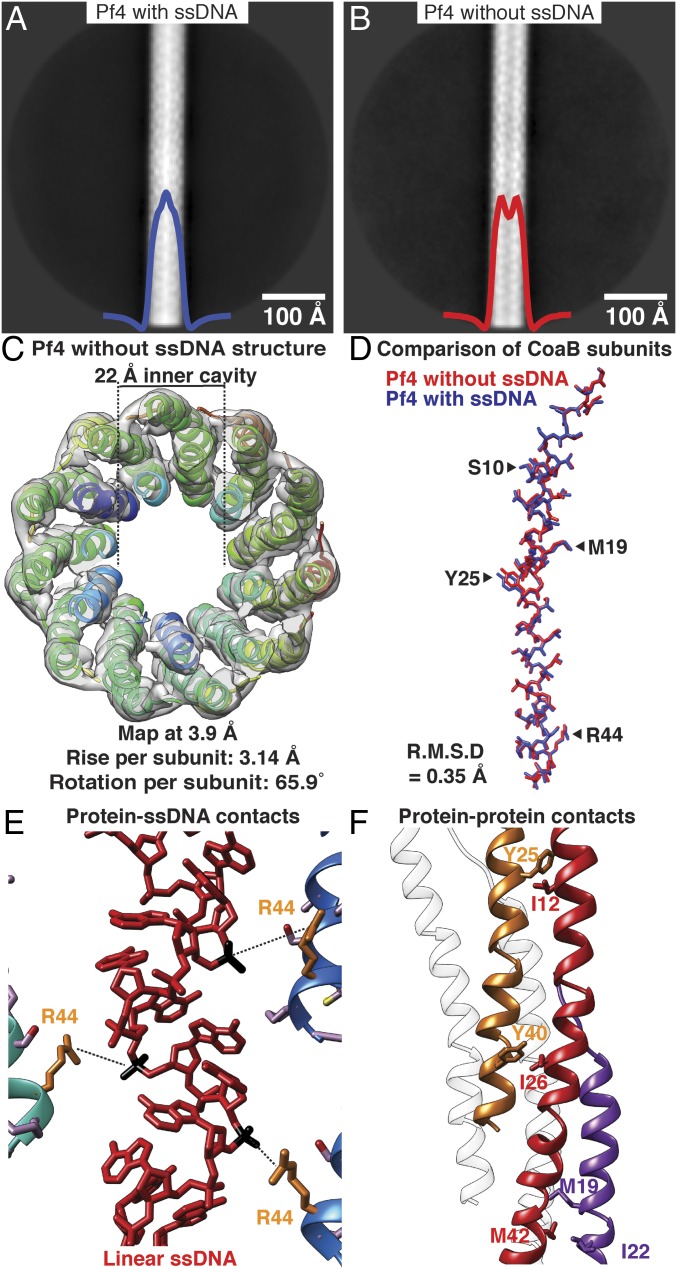
Fig. 2.
Cryo-EM structure of Pf4 filament without ssDNA at 3.9-Å resolution. (A) Two-dimensional class average of Pf4 with ssDNA, showing density in the core of the phage, indicated by peak in the horizontal density profile overlaid on average (blue curve). (B) Class average of Pf4 without ssDNA; a dip is observed in the horizontal density profile in the center of the average (red curve). (C) Top view of the cryo-EM structure of Pf4 without ssDNA at 3.9-Å resolution. Density is displayed as a gray isosurface and CoaB subunits as ribbons. The structure confirms lack of ssDNA in the 22-Å inner cavity. (D) Comparison of CoaB structure from Pf4 filaments with (blue) and without ssDNA (red) shows that the structures are almost identical (RMSD 0.35 Å, see Movie S2). (E) Magnified view of the CoaB:ssDNA interactions in the native Pf4 structure (from Fig. 1). Phosphates (black) of the linear ssDNA (red) are weakly coordinated by arginine 44 residues (orange) of CoaB (distance 5.4 Å). (F) Magnified view of CoaB:CoaB interactions within Pf4 filaments shows intersubunit hydrophobic interactions that stabilize the capsid.