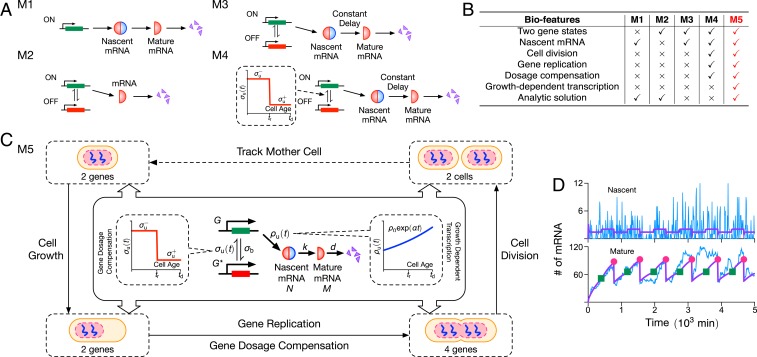
Fig. 1.
(A) Illustration of four models of stochastic mRNA dynamics in the literature. Note that nascent mRNA is shown as joined blue and red semicircles, illustrating its unspliced nature (blue for introns and red for exons), while mature mRNA being composed of only exons is shown as red semicircles. The models describe transcription (constitutive, e.g., M1; or intermittent, e.g., M2, M3, and M4), mRNA maturation (M1, M3, and M4), and details of the cell cycle (M4); their biological features are compared in B and discussed in the main text. Only the simplest of these two models (M1 and M2) have been analytically solved. The model (M5) proposed in this article is illustrated in C: It builds upon model M4 by adding growth-dependent transcription, describes maturation as a stochastic process, and has the major advantage of being analytically solvable. The model is composed of nonreactive components (dosage compensation, replication, cell division, and growth-dependent transcription) and reactive components; the latter are shown in the central boxes where denote genes in the ON, OFF states; is nascent mRNA; and is mature mRNA. (D) We show stochastic simulations of M5 using the SSA (25), where the purple lines denote the mean, and a typical time series is shown in blue. The green squares and red dots indicate the gene-replication time () and cell-division time (), respectively. We use parameters measured for Nanog in mouse embryonic stem cells from ref. 26: , , , and . The gene-replication time min, cell-division time min, maturation rate , and gene-dosage parameters are reported in ref. 19 for the same type of cells. Note that we set , meaning that there is no growth-dependent transcription. Each realization is initiated with zero nascent and mature mRNAs and the gene in the ON state.