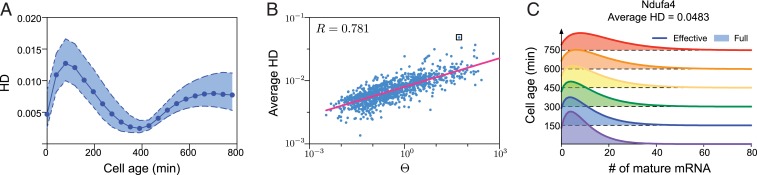
Fig. 4.
Effective negative binomial approximation for the mature mRNA number distribution of our model. The approximation is obtained by matching the cyclo-stationary mean and variance of our model to those of an effective negative binomial distribution. (A) We calculate the HD between the effective distribution and the full distribution for 21 equidistant time points through a whole cell cycle. Each point corresponds to the HD median for 1,051 genes in mouse embryonic stem cells (same data used for Fig. 3C), whereas the broken lines show the 25% and 75% quantiles. Note that the four parameters td = 780 min, , tr = 400 min, and are the same for all genes. (B) A plot of the HD for each gene averaged over 21 time points in the cell cycle versus the index ; the two quantities are linearly correlated in log space with Pearson correlation coefficient . (C) The matching of the effective and full distributions in time for the gene Ndufa4, which has the largest HD in B (shown as a boxed point).