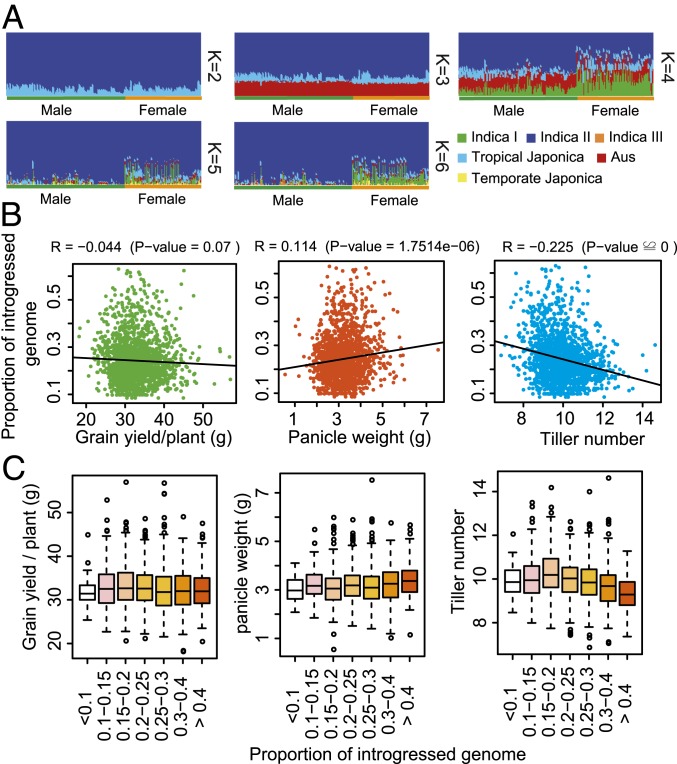
Fig. 2.
The heterotic level of hybrids increased with increasing exogenous genome introgression. (A) Population structure difference between male and female parents under different numbers of ancestor populations (K represents the prior number of ancestor populations). All 171 male and 104 female parents were integrated with ∼4,200 previously reported resequenced landraces and improved varieties to conduct the population structure analysis. The reasonable number of ancestor populations was determined using five-fold crossvalidation. Only SNPs on the 50k SNP chip were used to conduct the population structure analysis. Aus represents the south Asia origin aus rice subpopulation. (B) Correlation between degree of genome introgression and grain yield per plant (Left), panicle weight (Middle), and tiller number (Right) of hybrids. The effects of year, location, and population were regressed out using a general linear model for each trait, and the trait values for all hybrids in the study were used to conduct the analysis. (C) Comparison of hybrid yield traits with level of exogenous genome introgression. The yield traits included grain yield per plant (Left), panicle weight (Middle), and tiller number (Right). Hybrids were grouped by their cumulative levels of exogenous genome introgression (male parent + female parent). The bar in the middle of each box plot indicates the 50th quantile of the trait value for the group.