Fig. 1.
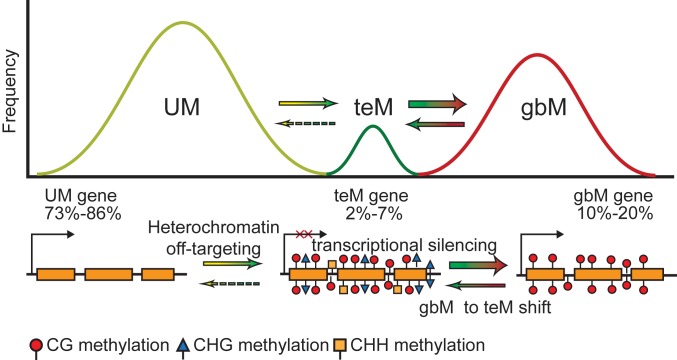
Genic DNA methylation patterns represent a continuum of epigenetic states. In A. thaliana, most genes located within euchromatin lack DNA methylation (UM). Some genes are prone to off-targeting by the heterochromatin machinery, which can result in transposable element-like DNA methylation (teM), characterized by cytosine methylation in all sequence contexts, CG, CHG, and CHH (H = A, C, or T), and transcriptional silencing. To combat the negative consequences of off-targeting of DNA methylation to genes, cells encode pathways that disrupt heterochromatin formation at genes by promoting the loss of non-CG methylation. This allows CG methylation to be maintained passively by maintenance methyltransferases, as CG methylation alone is inconsequential for transcription, which results in the strictly CG methylation pattern characteristic of gene body methylation (gbM). gbM then increases the probability of a gbM-to-teM shift due to feedback loops associated with DNA and histone methylation. The proportion of genes characterized by each methylation state within an individual vary due to factors that influence homeostatic targeting of DNA methylation. Percentages indicate the variation in the proportion of genes classified with each methylation pattern across 725 A. thaliana accessions.