Figure 4: Structure-based kinetic model for late steps in bacterial translation initiation.
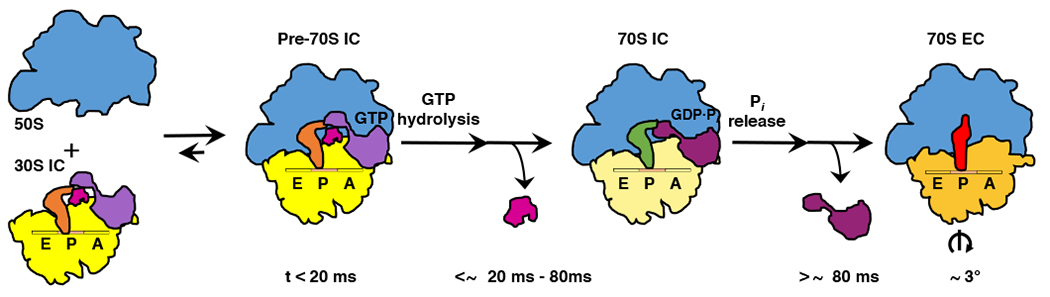
Cartoon depicting the timing of structural and molecular events during late steps in bacterial translation initiation. Within the first ~20 ms after mixing 50S subunits and 30S ICs, 50S subunits (blue) reversibly join to the majority of 30S ICs (yellow) to form transient Pre-70S ICs. Conversion of the majority of these Pre-70S ICs into 70S ICs takes place within ~20–80 ms after mixing of 50S subunits and 30S ICs and begins with the rapid hydrolysis of GTP on IF2 (light purple). GTP hydrolysis is followed by the dissociation of IF1 (magenta), repositioning of dIV of IF2 (dark purple), and formation of IF2-ribosome interactions and inter-subunit bridges that stabilize the ribosome in its semi-rotated inter-subunit orientation and the fMet-tRNAfMet in its 70S P/I configuration. Within the next several hundred milliseconds, the majority of 70S ICs mature into 70S ECs in a process that begins with release of Pi from IF2 and dissociation of the GDP-form of IF2 from the 70S IC, events that enable rotation of the ribosomal subunits into their non-rotated inter-subunit orientation, rearrangement of fMet-tRNAfMet into its P/P configuration, untangling of the 3’ CCA-fMet tail of fMet-tRNAfMet, and relocation of the fMet moiety of fMet-tRNAfMet into the PTC in preparation for formation of the first peptide bond upon delivery of the first aminoacyl-tRNA into the ribosomal aminoacyl-tRNA binding (A) site.
