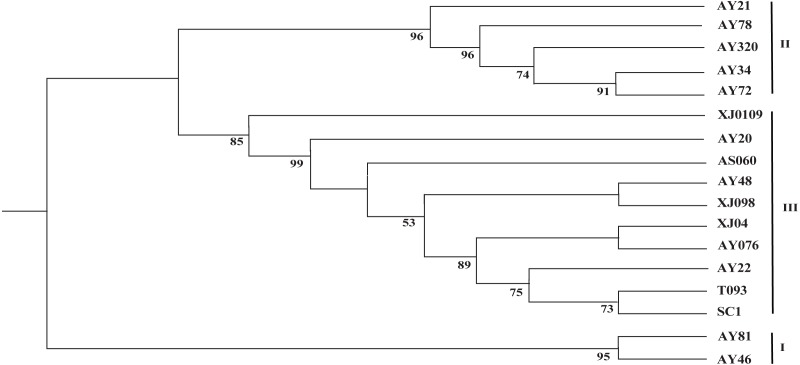
Figure 3. Phylogenetic tree constructed based on the complete chloroplast genomes of 17 Aegililops tauschii accessions by maximum likehood (ML) method.
Bootstrap support values (<50%) were hided. The phylogenetic tree resulting from analysis of 135,984 bp in the alignment length of chloroplast genomes with all gap positions removed, including long stretches of the same nucleotide, short sequences appearing in opposite orientation and some sequences consisting short repeats.