Abstract
In the mammalian genome, the clustered protocadherin (cPcdh) locus is a paradigm of stochastic gene expression with the potential to generate a unique cPcdh combination in every neuron. Here, we report a chromatin-based mechanism emerging during the transition from the naive to the primed states of cell pluripotency that reduces by orders of magnitude the combinatorial potential in the human cPcdh locus. This mechanism selectively increases the frequency of stochastic selection of a small subset of cPcdh genes after neuronal differentiation in monolayers, months-old organoids, and engrafted cells in the rat spinal cord. Signs of these frequent selections can be observed in the brain throughout fetal development and disappear after birth, unless there is a condition of delayed maturation such as Down Syndrome. We therefore propose that a pattern of limited cPcdh diversity is maintained while human neurons still retain fetal-like levels of maturation.
Editorial SUMMARY:
Short and long-term cultures of human stem cell-derived neurons reveal that a pattern of restricted selection of clustered protocadherin isoforms, pre-established in pluripotent cells, distinguishes immature from mature neurons.
Protocadherin (Pcdh) proteins are the largest subgroup of the cadherin superfamily of cell-adhesion molecules1. The clustered subtype (cPcdh) is encoded by 53 neuronal genes arranged in three adjacent clusters in the human genome (the α, β, and γ clusters)2–4. Forty-eight of these 53 genes are expressed such that every individual neuron expresses a small subset that is stochastically selected (PCDHA1-13 in the α-cluster, PCDHB1-16 in the β-cluster, and PCDHGA1-12 and PCDHGB1-7 in the γ cluster)2–4. This feature provides extraordinary cell-to-cell diversity with a combinatorial potential to express a ‘unique’ cPcdh selection in every neuron in the brain2–5. These selections mediate self/non-self-recognition through homophilic trans-interactions at the surface of neurons that contribute to the formation of neural networks6–10. The remaining five cPcdh genes (PCDHAC1-2 in the α-cluster and PCDHGC3-5 in the γ-cluster) are not expressed stochastically2–4.
The basic regulatory principles underlying the stochastic expression of the cPcdh isoforms were first established in mice11,12. In humans, cPcdh isoforms have been studied in neuroblastoma cell lines, in which each line consistently expresses a distinct cPcdh selection. In these cells, it has been determined that cPcdh selections are defined by a stochastic process of cPcdh promoter activation13 and by enhancers that determine their cell-type-specific regulation12,14–18. The stochastically selected promoter subset is recognizable by the accumulation of lysine 4 trimethylation on histone H3 (H3K4me3) and the binding of CCCTC-binding factor (CTCF) and cohesin-subunit Rad2115,19; the last two are organizers of the spatial configuration in the cPcdh locus12,14–18. In human neurons, however, the stochastic selection of cPcdh promoters remains to be investigated, although some studies have interestingly connected epigenetic (dys)regulation of the cPcdh locus to neurological and psychiatric disorders, Down syndrome, biological age, and histories of childhood abuse and prenatal alcohol exposure20. Here, we describe how our unrelated efforts to characterize neurons generated from single-cell-derived subpopulations of human induced pluripotent stem cells (hiPSCs) led us to uncover unexpected patterns of human cPcdh selections.
RESULTS
Limited cPcdh combinatorial potential in hiPSC-derived neurons.
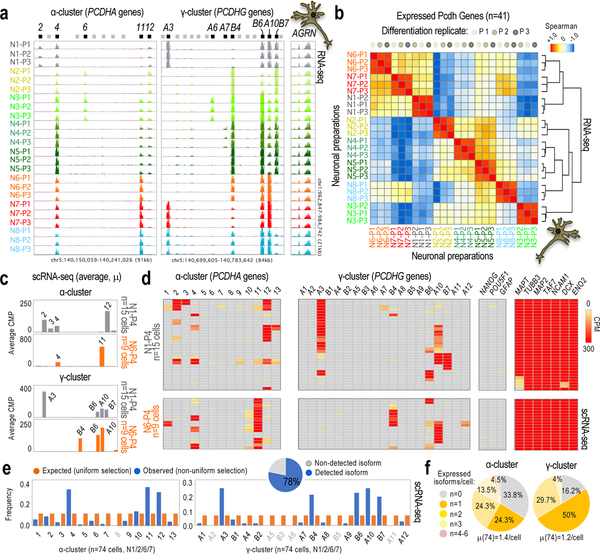
In previous studies, we applied genome editing to the Craig Venter-B (CVB) hiPSC line, which required a step of single-cell isolation and clonal expansion of edited and non-edited cells21,22. From eight of these single-cell-derived subpopulations (hiPSC1–8, Supplementary Table 1), we derived neurons (N1–8) in three rounds of neuronal differentiation (P1–3) using a different passage of neuronal progenitor cells (NPCs) in every round. After three weeks of differentiation, we applied fluorescence activated cell sorting (FACS) to enrich for similar subpopulations in every preparation. After a recovery period of 10 days, we profiled the n=24 final preparations by RNA-sequencing (RNA-seq; Supplementary Fig. 1a). Surprisingly, the eight subpopulations (N1–8) are distinguishable by their individual selection of expressed α/γ-cPcdh isoforms, regardless of the edited or non-edited genotypes and the round of differentiation. The selections, furthermore, show a preference to contain α/γ-cPcdh isoforms from only a small subset of the 48 options available; for example, some isoforms located towards the 3’ end of the γ-cluster (γB6, γA10, and γB7; Fig. 1a and Supplementary Fig. 1b,c). We corroborated the association of cPcdh selection to clonal origin by unsupervised clustering analysis (Fig. 1b), and also using alternative differentiation strategies (Supplemental Note and Supplementary Fig. 2a–d). Single-cell (sc)RNA-seq analysis further confirms an association of cPcdh expression to clonal origin (e.g. compare N1/6 in Fig. 1a,c), adding the important insight that each cPcdh selection represents a meta-signature of cumulative frequencies of smaller combinations, rather than repetitions of the same cPcdh selection in every cell (Fig. 1d and Supplementary Fig. 3a–c). We note that we detected 78% of the 48 possible, stochastically selected α/γ-cPcdh isoforms in the analysis of n=79 neurons (by scRNA-seq), with 19% (14) repeated α/γ combinations (Fig. 1e, pie chart, and Supplementary Fig. 3a). We suspect, therefore, that every α/γ-cPcdh isoform still has an opportunity for stochastic selection despite the strong clonal preferences (Fig. 1e,f). Together, these analyses suggest: (i) that hiPSC-derived neurons express signatures of α/γ-cPcdh isoforms with restricted variation that reduces the combinatorial potential in the cPcdh locus (a model of non-uniform probability of cPcdh selection); and, (ii) that this property is apparently established in progenitor hiPSCs before the stimulation of neuronal differentiation.
Fig. 1: Non-uniform probability of cPcdh selection in hiPSC-derived neurons.
a, RNA-seq data showing α/γ-cPcdh expression in three independent differentiation replicates (n=3, P1–P3) of neurons generated from NPCs derived from n=8 different single-cell-derived hiPSC subpopulations (hiPSC1–8). AGRN expression shown as reference. Expressed/non-expressed 5’ α/γ-cPcdh exons indicated (black and grey bars, respectively). Genomic coordinates: hg18. Scale: identical in all tracks. See some quantifications in Supplementary Fig. 1c. b, Hierarchical clustering (Spearman-rank correlation) and correlation matrix analyses based on expressed cPcdh genes in at least one neuronal preparation (n=41 out of 48) based on a (5’-exon-only signal). Analysis shows co-segregation of differentiation replicates in cPcdh expression. Color code: maximum (+1) to minimum similarity (−1). c,d, Expressed α/γ-cPcdh genes in n=15 single N1 cells and n=9 single N6 cells from a fourth differentiation replicate (P4). Data based on scRNA-seq (counts per million, or CPM). Data shown as an average of single cells (in c) or as individual cells (in d). Comprehensive heatmap shown in Supplementary Fig. 3a. Markers: pluripotency (NANOG and POU5F1), glial (GFAP), and neuronal (MAPT, TUBB3, NCAM1, DCX, and ENO2) genes. e, Observed (non-uniform) versus expected (uniform) distribution of random cPcdh selections in single hiPSC-derived neurons (combining n=15 N1, n=38 N2, n=9 N6, and n=12 N7 cells; scRNA-seq in Supplementary Fig. 3a). X2-test, P-value=2.2e-16. The pie graph represents the fraction of α/γ-cPcdh isoforms (out of n=48) detected in at least one single neuron (n=74 cells). f, Number and average (μ) of expressed α/γ-cPcdh isoforms by neuron (scRNA-seq, n=74 cells).
Chromatin-based restriction of cPcdh variation.
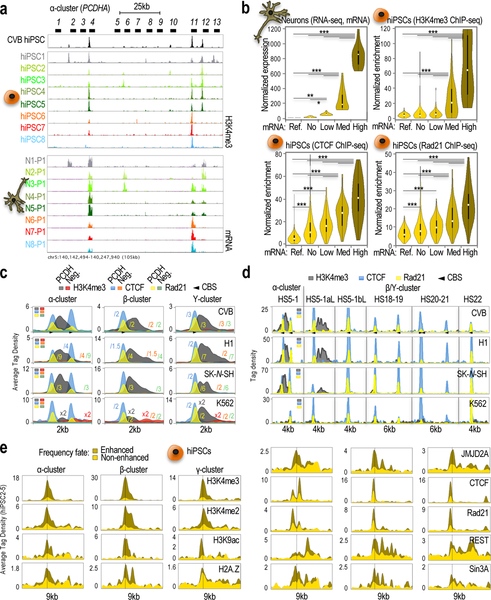
If a pattern of non-uniform probability of neuronal cPcdh selection is established in hiPSCs, we postulated that chromatin in these cells should exhibit evidence of this feature. In agreement, H3K4me3 analysis by chromatin immunoprecipitation followed by sequencing (ChIP-seq) in hiPSC1–8 reveals a pattern of H3K4me3 deposition along the cPcdh locus analogous to the pattern of α/γ-gene expression observed in clonally derived neurons (Fig. 2a and Supplemental Fig. 4a). The correlation (calculated with a Pearson’s coefficient of 0.72, P-value<0.0001) is corroborated in a side-by-side comparison of ChIP-seq and RNA-seq data stratified by levels of cPcdh expression (Fig. 2b, top panels). Likewise, CTCF and Rad21 ChIP-seq analyses reveal that CTCF/Rad21 loading in hiPSC1–8 correlates with expression in derived neurons, although not as strongly as H3K4me3 (Pearson’s coefficient=0.48, P-value<0.0001, Fig. 2b, bottom panels). These observations are not a byproduct of the editing process (previously applied to our cells) since hiPSC1 and hiPSC8, in particular, are cases of failed genome editing21,22; but also, because single-cell-derived hiPSC subpopulations without editing history, generated from a different line of the same donor (Craig-Venter I or CVI), also show clonal patterns of cPcdh selections (Supplementary Fig. 4b,c). Moreover, these observations are not an aberrant feature of the reprogramming process, since single-cell-derived subpopulations generated from blastocyst-derived human embryonic stem cells, or hESCs (in particular, the HUES 9 line), similarly show clonal patterns of cPcdh selections (HUES9 1.7–1.9; Supplementary Fig. 4d). We note that the parent CVB and CVI lines do share patterns of H3K4me3 accumulation distinct from the parent HUES9 line, although this feature is not attributable to hiPSC or donor identity (Fig. 2a and Supplementary Figs. 4a,b,d, e; other examples will be shown in Supplementary Fig. 7c; see also Supplementary Note and Supplementary Fig. 4f). On a meta-scale and by clusters, however, H3K4me3, CTCF, or Rad21 accumulation is virtually indistinguishable between hiPSCs and hESCs; it is similar to the accumulation observed in neuroblastoma SK-N-SH cells (which robustly express cPcdh genes14–18, as opposed to hiPSCs/hESCs); and, it is distinct from leukemia K562 cells (which do not express cPcdh genes14–18, Fig. 2c,d; see also Supplemental Note and Supplementary Fig. 5a,b).
Fig. 2: Chromatin in hiPSCs mirrors expression in hiPSC-derived neurons.
a, ChIP-seq data showing H3K4me3 accumulation along the α-cPcdh cluster in the parent CVB hiPSC line and the n=8 different single cell-derived sublines (hiPSC1–8) matched with RNA-seq data showing α-cPcdh expression in n=8 hiPSC1–8-derived neurons (N1–8 P1; data from Fig. 1a). Scales=set to maximum (ChIP-seq) or identical (RNA-seq); 5’ α-cPcdh exons and genomic coordinates (hg18) indicated. b, Violin plots of matching RNA-seq signal (top-left; n=3 differentiation replicates for N1–8, n=24), H3K4me3 ChIP-seq signal (top-right, one culture from each subline; hiPSC1–8, n=8), CTCF ChIP-seq signal (bottom-left; two independent cultures from each subline; hiPSC1–8, n=16), and Rad21 ChIP-seq signal (bottom-right; two independent cultures from each subline; hiPSC1–8, n=16) stratified by groups based on neuronal expression levels (top-left). Groups: non-neuronal genes (n=11; “Ref”); “non”-expressed cPcdhs (n=193 out of 384, or 48×8); “low”-expressed cPcdhs (n=72); “medium”-expressed cPcdhs (n=57); and, “high”-expressed cPcdhs (n=26). One-way ANOVA, multiple comparison test, P-value<0.01 (*), <0.001 (**), or <0.0001 (***). Shown median, interquartile range (25th and 75th). Error bars represent 95% confidence intervals. c,d, 2/4/6kb-wide meta-profiles of H3K4me3/CTCF/Rad21 ChIP-seq averaged tag density along cPcdh promoters by cluster (in c), or at distal regulatory sites (in d) in the indicated cultures (experiments as in b). “Neg” represents random coordinates. Scale adjustments indicated (e.g. x2 and /2 refer to signal multiplied/divided by 2, respectively). CTCF sites/orientation indicated. e, 9-kb-wide meta-profiles of ChIP-seq data for activating and repressive components (n=4 ChIP experiments [hiPSC2–5] for component). Promoters segregated by H3K4me3 enrichment/non-enrichment (enhanced/non-enhanced, respectively).
To gain further insights into the chromatin architecture defining the neuronal selection of cPcdh genes in hiPSCs/hESCs, we leveraged the rich repertoire of ChIP-seq data generated by the ENCODE Consortium, identifying at least n=23 datasets showing robust ChIP-seq signal across the cPcdh locus in hESCs (Supplemental Fig. 5d). These data show, for example, no H3K27me3 enrichment on cPcdh promoters, thus against a model of ‘bivalent domains’ (or, H3K4me3/H3K27me3 co-enrichment23) in which H3K27me3 counteracts H3K4me3-activating actions; although alternative repressors could be found on these promoters (Supplemental Fig. 5d). We therefore used our panel of single-cell-derived hiPSC sublines (in particular hiPSC2–5) to formally test whether α/γ-cPcdh promoters accumulating H3K4me3 (hereafter referred to as ‘enhanced’ promoters) also recruit repressors, which would explain the combination of inactive transcriptional state with a subsequent fate of higher frequency of neuronal selection. In particular, we profiled the RE1-silencing transcription factor (REST), its corepressor SIN3A, and the associated lysine demethylase Jumonji domain-containing protein 2A (JMJD2A/KDM4A)15,19. In addition, we profiled activating H3K4me2, H3K9ac, and histone variant H2A.Z. Segregated by fate of frequency selection (‘enhanced’ and ‘non-enhanced’; the latter corresponding to poorly/non-H3K4me3-enriched promoters), these analyses reveal that enhanced α/γ-promoters distinctively accumulate both activating and repressive chromatin compared to non-enhanced promoters (Fig. 2e). Together, these findings reveal a chromatin configuration in pluripotent stem cells that distinguishes the promoter subset with a fate of preferential neuronal selection, thus supporting the model that in vitro-generated neurons inherit the pattern of preferential cPcdh selections.
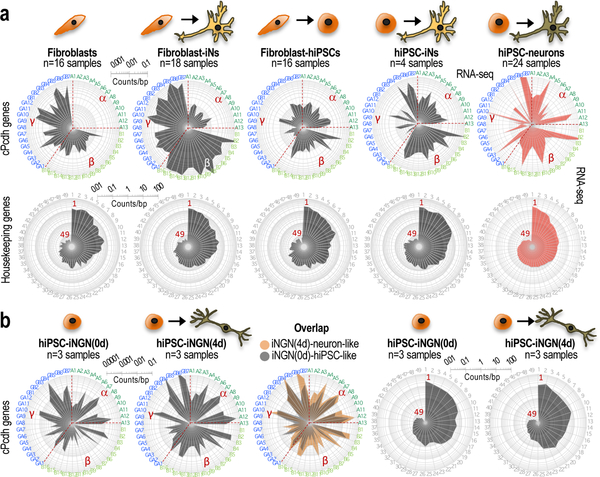
We further corroborated the conclusion that human neurons inherit cPcdh-locus features from non-neuronal progenitor cells using neurons derived from somatic cells without undergoing an intermediate step of cell pluripotency, also known as induced neurons or iNs24. These cells express cPcdh genes following the organizational properties of their source of somatic cells (Fig. 3a,b; see also Supplementary Note and Supplementary Fig. 6).
Fig. 3: In vitro-generated neurons inherit cPcdh-locus features from non-neuronal cells.
a, Log10-scale radar plots showing averaged RNA-seq data for the n=48 stochastically selected cPcdh genes (top) and n=49 housekeeping genes (bottom) in the indicated neuronal preparations (iNs or hiPSC-derived neurons) and progenitor somatic or pluripotent cells (skin fibroblasts or hiPSCs, respectively). Number of independent cultures indicated on top of each plot. See a detailed description in Supplementary Note. Data source: this study (right plots) and E-MTAB-3037 (the rest). Clockwise, cPcdh genes are shown in numerical and alphabetical order by clusters (color-coded in the periphery). b, Log10-scale radar plots showing averaged RNA-seq data for the n=48 stochastically selected cPcdh genes (the three left plots) and n=49 housekeeping genes (the two right plots), as in a. iNGN refers to iNs generated by neurogenin overexpression in hiPSCs, which is an alternative protocol of direct neuronal reprogramming to the protocol applied to derive iNs in a (data source: GSE60548). The middle plot is an overlap of the two radar plots shown on the left side. The number of independent cell cultures is indicated on top of each panel. See a more detailed description of these experiments in Supplementary Note.
Pre-setting cPcdh frequencies parallels the acquisition of the primed state.
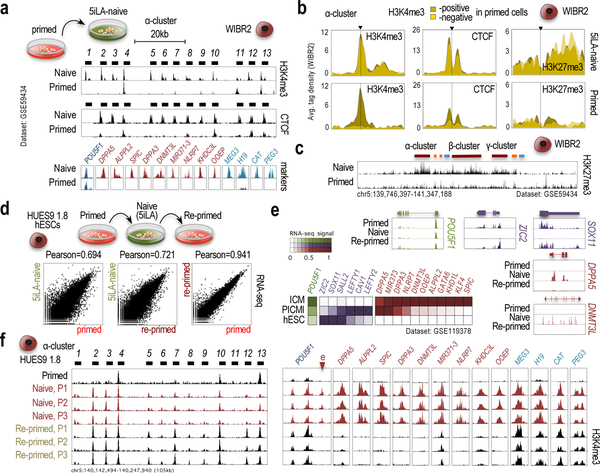
We next interrogated the timing of cPcdh-frequency ‘pre-setting’ in pluripotent stem cells. Some clues surface in the analysis of the hTERT-immortalized human fibroblast-like secondary reprogramming system (Supplementary Fig. 6b25). In this system, H3K4me3 accumulates on the cPcdh locus at the same time as it accumulates on promoters of pluripotency regulators (NANOG, LIN28A, and POU5F1), and on promoters of genes expressed in the preimplantation embryo (DPPA3, DNMT3L, MIR371–3, and NLRP726). Moreover, it is well established that hiPSCs/hESCs spontaneously progress from a preimplantation-like to a postimplantation-like stage during derivation, also known as naive and primed states, respectively27,28, which is accompanied by H3K4me3 removal from the promoters of preimplantation/naive genes25 (Supplementary Fig. 6b). Thus, we postulated that the pre-setting of cPcdh frequencies might occur during the process of naive-to-primed conversion. In agreement, primed hESCs subjected to culturing conditions that capture the naive state (in particular, 5iLA conditions or the 4iLA/6iLA variations Ref23,30), stimulate H3K4me3 and CTCF accumulation on most, rather than on only a small subset of cPcdh promoters (Fig. 4a). Segregating by enhanced and non-enhanced promoters, the patterns of H3K4me3/CTCF accumulation are virtually indistinguishable between these two classes of primed promoters after induction of the 5iLA-naive state (Fig. 4b and Supplementary Fig. 7a). And, further in support of a change in chromatin configuration, all cPcdh promoters accumulate H3K27me3 in 5iLA-naive cells (bivalent domains, Fig. 4c), in contrast to repressive H3K9me3, which accumulates on cPcdh promoters regardless of the 5iLA-naive or primed states (Supplementary Fig. 7b). In contrast, “naive”-inducing protocols unable to reactivate archetypical preimplantation markers do not alter the primed pattern of H3K4me3 accumulation on cPcdh promoters (Supplementary Fig. 7c), thus suggesting that only a naive state that recapitulates the preimplantation-like stage (e.g. 5iLA conditions29,30) replicates a state that ‘precedes’ the segregation of enhanced/non-enhanced cPcdh promoters.
Fig. 4: Differences in cPcdh-locus chromatin organization between naive and primed cells.
a, ChIP-seq data showing H3K4me3 and CTCF accumulation along the α-cluster (top/center panels; 5’-cPcdh exons indicated) and ChIP-seq data showing H3K4me3 accumulation on promoters regulating pluripotency (in blue), preimplantation (in red), and imprinted (in green) genes in 6iLA-naive and primed WIBR2 hESCs. Data source: GSE59434 and GSE69646. b, 9kb-wide meta-profiles of H3K4me3, CTCF, and H3K27me3 ChIP-seq signal on the n=13 α-cPcdh promoters in naive and primed WIBR2 hESCs. Promoters segregated by H3K4me3 enrichment in primed cells: H3K4me3-enriched (positive) or non- H3K4me3-enriched (negative). Data source: GSE59434 and GSE69646. c, H3K27me3 ChIP-seq signal along the cPcdh locus in 6iLA-naive and primed WIBR2 hESCs. Data source: GSE59434. d, Full transcriptome-wide comparisons of expression profiles (RNA-seq) between a culture from each of the listed pluripotency states (primed, 5iLA-naive, and re-primed) in HUES9 1.8 cells. Pearson correlation of the comparison (Refseq genes) indicated on top. e, RNA-seq tracks of relevant genes in the data shown in d. Pluripotency (in green), preimplantation (in red), and postimplantation (in blue) genes. Heatmap of RNA-seq signal of three independent cultures (each) of human ICM embryos, PICMI, and hESCs (data source: GSE119378). f, H3K4me3 ChIP-seq signal along the α-cluster and on preimplantation and imprinted promoters in primed (n=1), naive (n=3), and re-primed (n=3) single-cell-derived HUES9 1.8 hESCs. 5’ cPcdh exons indicated on top. Markers: pluripotency (in black, POU5F1 promoter), preimplantation (in red, including an enhancer [e] in the POU5F1 locus); and imprinted promoters (in green). Genomic coordinates (hg18).
If the reversion of the 5iLA-naive state returns the cPcdh locus to a state that precedes the segregation of enhanced/non-enhanced promoters, returning it back to the primed state (or, ‘re-priming’) may generate a new set of cPcdh promoter selections different from those observed in the original primed version. To test this hypothesis, we exposed one of our single-cell-derived HUES9 sublines (HUES9 1.8) to the 5iLA protocol and returned it to the primed state (Fig. 4d). First, we corroborated that the primed and re-primed states are remarkably similar at a transcriptome-wide scale (Pearson’s coefficient=0.941) and differ from the naive state to similar extents (Pearson’s coefficient=0.721 and 0.694, respectively; Fig. 4d and Supplementary Fig. 8a). Second, we corroborated that a panel of preimplantation genes expressed in the inner cell mass (ICM) of the human blastocyst is expressed in naive HUES9 1.8 cells (in cayenne in Fig. 4e and Supplementary Fig. 8b), whereas postimplantation genes expressed shortly after ICM-blastocyst derivation (post-ICM intermediate stage or PICMI27,28) are expressed in primed and re-primed HUES9 1.8 cells (in purple in Fig. 4e and Supplementary Fig. 8b). Despite this successful process of re-priming, the cPcdh locus does not recover the original primed configuration, indicating that resetting occurred without memory of the original primed configuration (Fig. 4f and Supplementary Fig. 8c; see also Supplementary Note and Supplementary Fig. 9). We note that a second feature that did not recover the original primed configuration is the chromatin organization on promoters of some imprinted genes (see MEG3, H19, CAT, and PEG3 panels in Fig. 4f and Supplementary Fig. 9,10). Together, we conclude that the pre-setting of frequencies of cPcdh selection occurs during the naive-to-primed conversion, and that reversion to a naive state that activates archetypical pre-implantation-like markers resets these selections.
Restricted cPcdh selections in mouse primed cells.
Unlike hESCs and hiPSCs, mouse (m)ESCs and iPSCs are regarded as naive cells after derivation31. Similar to human 5iLA-naive cells, therefore, H3K4me3 and CTCF accumulate on every α/γ-cPcdh promoter in mESCs and iPSCs, as does H3K27me3, as previously noted23 (Supplementary Fig. 11a, mESCs and iPSCs, and Supplementary Fig. 11b). During derivation of mouse iPSCs, H3K4me3 and H3K27me3 accumulate on α/γ-cPcdh promoters and, at the same time, on promoters of pluripotency and pre-implantation genes as observed in human cells (Supplementary Figs. 6b and 12). Primed cPcdh configurations can be observed in mouse postimplantation(-like) cells derived from E5.5–6.5 embryos or from mESCs differentiated into E7.5-like cells, which are known as epiblast-derived stem cells, or EpiSCs (Supplementary Fig. 11a, EpiSCs, H3K4me3/H3K27me3). We found one exception, which is mESCs differentiated into E5.5-like cells, which are known as epiblast-like cells, or EpiLCs. These cells resemble naive mESCs with regard to the organization of the cPcdh locus, thus being analogous to E5.0 rather than to E5.5 embryos, or an incomplete E5.5 conversion (Supplementary Fig. 11a, mESCd-E5.5). Perhaps in support, EpiLCs show Dppa5 activation (preimplantation gene), in contrast to mouse E5.5 embryos (Supplementary Fig. 11a, mESCd-E5.5 and E5.5). We therefore propose that the adoption of mouse primed/restricted cPcdh configurations occur at a stage developmentally analogous to peri-implantation (E5.0-E5.5). Finally, we successfully corroborated that mouse naive and primed cells lack and exhibit, respectively, patterns of already made cPcdh selections in a panel of single-cell-derived mESC and EpiSC subpopulations (Supplementary Fig. 13a–c). In sum, analysis of mouse data suggests that the pre-setting of restricted cPcdh-frequency selections occurs in mouse cells as in human cells (and it might occur also in other species; Supplementary Fig. 14).
Having found parallels between the configuration of the cPcdh locus between human and mouse pluripotent cells, we next sought to examine the cPcdh locus beyond the postimplantation state in the developing mouse embryo. In the γ-cluster, surprisingly, a bivalent (i.e. preimplantation-like) state returns to every γ-promoter at around E8.5, in parallel with Pou5f1 inactivation and the onset of organogenesis; H3K27me3 is later gradually eliminated, fully disappearing after birth (Supplementary Fig. 15). In the α-cluster, in contrast, H3K4me3 (without H3K27me3) accumulates on every α-promoter after E13.5 (Supplementary Fig. 15). Thus, the ‘active’ state (defines as H3K4me3-positive, H3K27me3-negative promoters) would be established earlier in the α-cluster than in the γ-cluster during mouse brain development. We therefore propose that, although signs of the pre-setting of restricted cPcdh-frequency selections can be observed in early mouse embryos (before E8.5) and in primed cells, mouse neurons (as opposed to human neurons in vitro) would not inherit these selections that seem to disappear during development/differentiation32,33.
Stability of the hESC/hiPSC-defined cPcdh configurations.
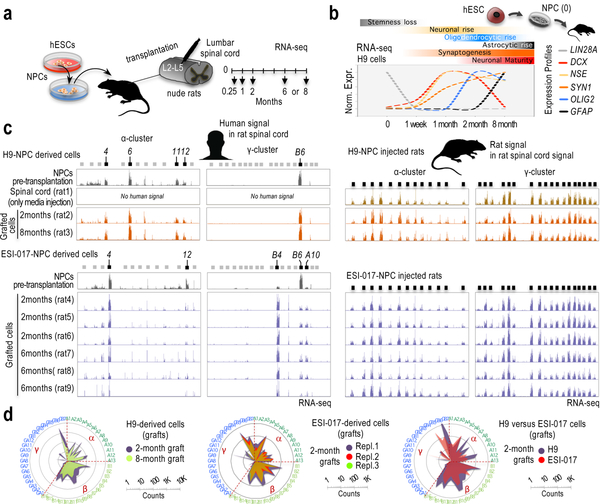
Since in vitro-generated human neurons are considered to be immature neuronal cells, we next interrogated whether these cells must reach a certain level of maturation (or differentiation) to erase or override the inherited frequencies of restricted cPcdh selections. We applied a recent protocol to generate cortical organoids that stimulates oscillatory network dynamics observed in the brain of preterm neonates34. In support of substantial ‘maturation’, 10-month-old cortical organoids do not express the early post-mitotic marker DCX35 and express astrocytic genes (GFAP and S100B)36, in contrast to our monolayered cultures (Supplementary Fig. 16a). These organoids also express PCDHGC5 and suppress PCDHGC4, a property observed in mouse and rat brains around birth or soon after37–39 (Supplementary Fig. 16b). Still, cPcdh expression shows signs of restricted variations (Supplementary Fig. 16c). As a second strategy to stimulate neuronal maturation, we injected H9 and ESI-017 hESC-derived NPC suspensions in the central grey matter of the rat spinal lumbar region40. After 1 week, 1, 2, or 8 months engrafted in H9-NPC-injected rats, and 2 or 6 months engrafted in ESI-017-NPC-injected rats, cells were processed for immunostaining and RNA-seq analyses (Fig. 5a). While these analyses indicate certain level of neuronal maturation (Supplemental Note and Supplementary Figs. 17,18), human grafts still retain patterns of restricted cPcdh configuration, in contrast to the surrounding, adult rat tissue (Fig. 5c,d).
Fig. 5: Signs of hESC-guided cPcdh signatures are remarkably stable in vitro and in vivo.
a, Experimental scheme. b, Relative and normalized RNA-seq signal in H9-derived NPCs prior transplantation (“0”, n=2 samples) and in grafted cells after transplantation, 1 week or 1/2/8 months, as indicated (one rat at each time point). Profiles normalized to 1 (maximum expression for each gene). RNA-seq values and additional genes are shown in Supplementary Fig. 18a. Markers: LIN28A (NPC identity); ENO2 (neuronal identity); DCX (early postmitotic neurons); SYN1 (synaptogenesis); GFAP (astrocytic identity); and, OLIG2 (oligodendrocytic identity). c, RNA-seq signal along the human α-cluster (first column) and γ-cluster (second column), and along the rat α-cluster (third column) and γ-cluster (fourth column) in H9-NPC engrafted cells and surrounding rat cells in the spinal cord, respectively (top) and ESI-017-NPC engrafted cells and surrounding rat cells in the spinal cord, respectively (bottom). Rat and human reads were computationally separated and independently aligned to the rat and human genomes, respectively. Samples: hESC-derived NPCs prior transplantation (n=2), media-only-injected spinal cord (n=1), and 2, 6 or 8-month engrafted cells, as indicated (a rat at each time point in H9-NPC injected cases and n=3 rats at each time point in ESI-017-NPC injected cases). 5’ cPcdh exons indicated on top; expressed exons highlighted in black. Y-axis is adjusted to max in each track. d, Log10-scale radar plots of averaged RNA-seq signal for the n=48 stochastically selected cPcdh genes in the indicated conditions. (clockwise, in numerical/alphabetical order by clusters, color-coded in the periphery).
Signs of restricted cPcdh choices in fetal brains.
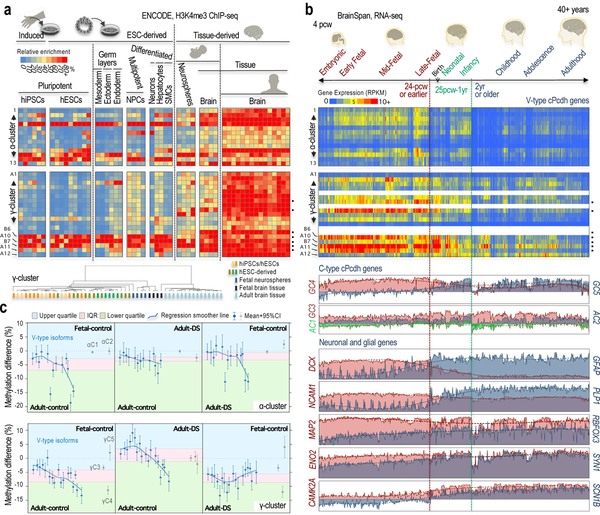
It is possible that a pattern of restricted variation in the selection of α/γ-cPcdh isoforms could be an exclusive characteristic of laboratory-generated human neurons. We therefore examined the potential presence of this feature in postmortem human fetal and adult brains (Fig. 6a and Supplementary Fig. 19a,b). In the γ-cluster, H3K4me3 follows the expected pattern of relatively uniform selection in adult brains, but not in fetal brains. We computed the range (or variance, σ2) of H3K4me3 accumulation, which corroborated a difference between the relatively uniform, adult tissue and the relatively non-uniform, fetal tissue (P-value=0.005), as well as between the former and hiPSC/hESCs, hESCs-derived cells, and fetal neurospheres (the latter are NPC-enriched populations derived from fetal brains; Fig. 6a and Supplementary Fig. 19a). Unsupervised clustering analysis also segregates the distribution of H3K4me3 accumulation in adult brains apart from fetal brains and in vitro samples (Fig. 6a, dendrogram). Importantly, furthermore, fetal tissue and our laboratory-generated cells share the pattern of higher H3K4me3 enrichment on the promoters of some γ isoforms (γB6, γA10, γB7, and γGA11; Fig. 6a, γ-cluster heatmap). In the α-cluster, in contrast, the range of H3K4me3 distribution is only slightly different between fetal and adult tissue (P-value=0.098), or between fetal tissue and neurospheres (P-value=0.073), although it is statistically different between adult brains and fetal neurospheres (P-value=0.011; Fig. 6a, α-cluster heatmap). Unsupervised clustering analysis co-segregates fetal and adult brains, and fetal neurospheres segregate with in vitro cultures (Supplementary Fig. 19c). We note nonetheless that the α-cluster, even in the adult brain, exhibits some signs of preferential H3K4me3 enrichment (towards the 5’ and 3’ ends), as recently observed in mESC-derived neurons33 and in mouse neurons41 (Fig. 6a, α-cluster; Supplementary Fig. 19d, α-cluster, brain).
Fig. 6: Two distinct types of cPcdh diversity distinguish fetal and adult brain tissues.
a, Heatmap, H3K4me3 ChIP-seq signal on α/γ-cPcdh promoters relative to the highest enrichment observed in each sample (as a percentage over max, or 100%). Sample scheme on top, samples listed in Supplementary Fig. 19a. Postmortem human brains: fetal, n=2 donors, gestational stage=17-pcw, or post-conception weeks; fetal, germinal matrix, n=2 donors, 20-pcw; and, adult brain, n=3 donors, range=73–81 year-old, n=7 independent samples. Neurospheres from postmortem fetal brain, n=4 donors, range=15–17-pcw. The most H3K4me3-enriched γ-promoters in human fetal brains are indicated (black dots). Dendrogram, hierarchical clustering analysis of the same samples shown in the heatmaps; one minus Spearman-rank correlation. Samples color-coded by source. b, Heatmap, RNA-seq signal (RPKM, exonic, BrainSpan cohort) of α/γ-cPcdh genes in the n=524 postmortem samples of the human developing and adult brain (ranging from 4-pcw to 40+ years of age and a variety of different brain regions42). Data not available for four γ-isoforms (in blank). Profiles, RNA-seq signal (RPKM, BrainSpan) for non-stochastically selected cPcdhs and neuronal and glial markers. The most H3K4me3-enriched γ-promoters in fetal brains are highlighted (black dots). c, Mean differential methylation with 95% confidence intervals (CI) on stochastically selected (blue circles) and non-stochastically selected (grey circles) α/γ-promoters (top/bottom panels) in postmortem brain samples of control and Down syndrome subjects, as indicated in the upper/lower side of each panel (sample numbers and additional labeling provided in Supplementary Fig. 21; Supplemental Note). Background colored based on interquartile range (IQR, pink) and upper/lower quartiles (blue/green) of stochastically selected promoters.
Next, we examined cPcdh expression in the Allen BrainSpan collection, which is based on n=524 postmortem tissue samples ranging from 4-pcw to 40+ years of age and a variety of different brain regions42 (Fig. 6b and Supplementary Fig. 20). In this dataset, PCDHGC437–39 and DCX43 reach the lowest expression after 1–2 years of age, whereas PCDHGC5 and synaptic and glial components (CAMK2A, SCN1B, PLP1, and GFAP42) reach the highest expression after this age (Fig. 6b, profiles). In the γ-cluster, the fetal expression of stochastically selected cPcdh genes follows a pattern analogous to that of fetal H3K4me3 accumulation (the latter shown in Fig. 6a), at least until two years of age (Fig. 6b, heatmap, γ-cluster); after that, cPcdh expression declines below the threshold of detection in most cases. Notably, the most highly expressed γ-isoforms have some of the highest levels of H3K4me3-promoter accumulation in fetal tissue and in hiPSC/hESC-derived cells (Fig. 6b, heatmap, e.g. promoters regulating the γB6, γA10, γB7, and γGA11 isoforms). In combination, ENCODE and BrainSpan data suggest that γ-isoforms are expressed in fetal brains with signs of preferential frequencies of selection, similar to those observed in laboratory-generated cells, although not as extreme, raising the possibility of some culturing effects. The fact that laboratory-generated cells show a fetal-like pattern of γ-cPcdh selection seems consistent with the general view that these cells represent fetal-like entities. In the α-cluster, the signal in the BrainSpan data is close or below to the limits of detection in most cases, although some 5’/3’ H3K4me3-enrichment can be observed starting shortly after 4-pcw (Fig. 6b, heatmap, α-cluster). The situation in the α-cluster is, therefore, less clear than in the γ-cluster, but if the α-cluster matures earlier than the γ-cluster in humans, as it appears that may occur in mouse embryos, perhaps this early stage is not represented in the BrainSpan cohort.
Fetal-like features in the γ-cluster of adult Down Syndrome brains.
Finally, we postulated that if a pattern of more or less restricted diversity of γ-cPcdh selections is a fetal(-like) feature, humans with temporal disturbances leading to delayed brain maturation, such as Down Syndrome, might aberrantly retain this feature in adult brains. To test this hypothesis, we leveraged a series of DNA methylation measurements in postmortem samples of frontal cortex brains44,45 (correlation of promoter methylation and cPcdh expression was previously validated in these samples44). In line with our hypothesis, a side-by-side comparison of fetal control and adult Down syndrome brains reveals that adult Down syndrome brains retain a distinct pattern of fetal hypomethylation towards the 3’ end of the γ-cluster, which is predicted to be associated with relatively high expression (Fig. 6c, bottom-right). In support, a side-by-side comparison of adult Down syndrome and control brains resembles a side-by-side comparison of fetal and adult control brains with regard to this fetal feature, despite a median age similarity of 45 versus 49.5 years in the adult Down syndrome/control comparison (Fig. 6c, bottom-center; compare to bottom-left). The fetal pattern appears enhanced in Down syndrome brains (Supplementary Fig. 21, right panel; see Supplemental Note). Together, these analyses suggest a retention of a fetal-like feature in the γ-cluster of adult Down syndrome brains, potentially in agreement with a model of delayed brain maturation in Down syndrome. Notably, this retention seems specific to the subset of stochastically selected cPcdh promoters, as the subset of developmentally regulated cPcdh promoters (PCDHGC4/C5) shows the expected patterns of DNA methylation associated with fetal and adult expression, respectively, in brain tissue (Fig. 6c).
DISCUSION
The stochastic expression of cPcdh isoforms was first reported in mouse neurons in 200546. Since then, multiple studies have expanded this finding2–4; and, today, the possibility that every mouse and human neuron expresses a unique combination of cPcdh isoforms is an attractive model to explain vast neuron diversity2,32. We are not aware, however, of any study that has examined yet this model in human cells. Here, we provide data indicating that the stochastic selection of cPcdh isoforms is more complex and dynamic than anticipated in human cells in light of previous work based on mouse neurons. First, we describe a new type of stochastic cPcdh-expression pattern characterized by unequal probability of α/γ-cPcdh selections that decreases the combinatorial potential in the cPcdh locus and increases the likelihood of cPcdh-signature repetitions among human cells. Second, we propose that this pattern is specific to fetal and fetal-like neurons (in the case of laboratory-generated cells), unless there is a condition of delayed brain maturation or aberrant maturational constraint that prolongs the immature version into the adult stage (for instance, we propose, in Down syndrome). And, third, we report that progenitor pluripotent cells pre-set this type of neuronal cPcdh-expression pattern when exiting the naive state, at a stage likely developmentally analogous to peri-implantation (mouse E5.0-E5.5 and human day 8–12), which entails the so-called ‘formative’ state of pluripotency47 (Supplemental Note). A corollary of this third finding is that, since human pluripotent cells spontaneously exit the naive state during in vitro derivation (in ICM outgrowths for hESCs27,28 and in the final stages of colony formation for hiPSCs25), hESC/hiPSC cultures are stable mosaics of cells with pre-set frequencies of neuronal α/γ-cPcdh selections. This property should be taken into consideration when isolating single cells from hESC/hiPSC cultures prior to inducing neuronal differentiation (e.g. when applying genome editing), since it will unintentionally isolate cPcdh-frequency pre-selections unless cell isolation precedes the loss of the naive state, e.g. under 5iLA conditions29,30 (used in this study) and likely also under T2iLGö conditions48. For this reason, we suspect, cells derived from two halves of a late ICM outgrowth (PICMI cells becoming hESCs)27,28 show different preset cPcdh configurations (see Supplementary Fig. 7c, WIBR1–3). We note that cPcdh configurations could be used as markers for hESC/hiPSC line or subline authentication purposes.
In mouse embryos, two previous studies also reported an epigenetic event across the cPcdh locus before the emergence of neuronal cells41,49; in particular, a process of de novo DNA methylation that is permissive of stochastic selections of cPcdh isoforms in adult Purkinje cells41. We suspect that this process could be connected to our observation of erasure in mouse cells, at around E8.5, of the cPcdh frequencies preset during the acquisition of the primed state. In humans, however, this early embryonic event might not exist, at least in the γ-cluster. We observed general hypermethylation of the cPcdh locus in adult brains compared to fetal brains (Fig. 6c), whereas the study in mice reported no major changes in DNA-methylation levels after E9.5 compared to adult brain41. We therefore predict that a mechanism of fetal cPcdh-signature erasure would occur in humans after birth (at least in the γ-cluster), which would be similar to what has been described for some retina genes in mice49. Otherwise, the pattern of cPcdh selections that we report cannot explain the extraordinary combinatorial potential presumed in the adult human brain. An important caveat of this model is that we have been unable to replicate the onset of the adult pattern of human cPcdh selection in laboratory-generated cells, which is perhaps expected considering that these cells only recapitulate a fetal stage or -at most- oscillatory network dynamics of preterm babies Ref.34. We therefore propose that the adoption of a pattern of adult cPcdh selections is a postnatal developmental milestone in the human brain whose recapitulation should be an aim of the development of cerebral organoid-based models. This milestone would be subsequent to PCDHGC4 silencing and PCDHGC5 activation, and subsequent also to expression of any other gene marker of neuronal maturation examined here. Due also to the current inability to recapitulate the adult stage of a human neuron in the laboratory, we have been unable to address the overarching question of whether having two types of cPcdh diversity, one fetal-one adult (being one more diverse than the other), has a functional relevance in human cells. We predict that increasing the likelihood of cPcdh-mediated matching (or, repetition) among fetal neurons will impact the processes of self/nonself-recognition, dendritic branching, and axonal tiling on a network scale6–10,50–55. In mice, for example, high likelihood of cPcdh matching is required for axonal tiling and assembly of serotonergic circuitries56. Further, mice genetically modified to increase the likelihood of cPcdh matching present abnormalities in higher cognitive functions in adult animals, interestingly, without grossly affecting fetal brain development57, and show higher levels of dendrite arborization9. On the contrary, low likelihood of cPcdh matching is required for proper axon convergence in the developing mouse olfactory system after birth10. Together, these studies may provide clues to the functional consequences of the developmental dynamics of cPcdh selection pre-setting and erasure.
In conclusion, we propose the exciting possibility that having two modes of cPcdh selection may play a role in the orchestrated establishment of connectivity in the human brain. Importantly, furthermore, we propose that the fetal model is pre-set by non-neuronal progenitor cells, perhaps as a form of maturational constraint that is only eliminated when the nervous system reaches certain age after birth. It will be, therefore, interestingly to explore how failing to eliminate this constraint may contribute to Down syndrome (although our findings in Down syndrome brains should be first replicated in a larger cohort and at the level of chromatin or expression) or, potentially, how it may contribute to other neurological and psychiatric disorders20. Our findings, furthermore, raise the interesting possibility that this constraint may affect the integration of hiPSC/hESC-derived cells into an adult nervous system for regenerative purposes.
METHODS
Human/mouse pluripotent stem lines and processes of single-cell isolation and expansion
We have complied with all relevant ethical regulations with regard to the use of pluripotent stem cells. The CV-hiPSC-B (or, CVB; RRID:CVCL_1N86, GM25430), and CV-hiPSC-I (or, CVI) lines were previously established58,59. Details about their generation, karyotypes, and pluripotency capacity were previously reported58,59. The eight genome-edited sublines derived from the parent CVB line were also previously reported21,22, but we have changed their names to hiPSC1–8 for convenience (matching between the original and the here-assigned names can be found in Supplemental Table S1). Each of the these sublines were generated by expanding individual hiPSC colonies after diluting 10,000 cells into a 10 cm plate pre-seeded with γ-irradiated mouse embryonic fibroblasts (MEFs; in-home generated). Five to seven days after plating, colonies were manually picked, transferred into 96-well plates pre-seeded with γ-irradiated MEFs, genotyped, and individually expanded21,22. The three single-cell-derived CVB and CVI sublines that did not undergo a process of genome editing were also previously generated from single cells separated by cell sorter21,22. Their original names have been maintained (CVB 1.7, CVB 1.8, CVB 1.9, CVI 1.13, CVI 1.14, and CVI 1.15; Supplemental Table S1). Before sorting, parent CVB and CVI lines were labeled for the cell surface marker Tra1–81 (BD Biosciences), and sorted Tra1–81-positive cells (BD FACSARIA cell sorter) were seeded in individual wells of a 96-well plate pre-seeded with γ-irradiated MEFs. These cells were maintained and passaged as previously reported21,22. At passage five, cells were labeled with Tra1–81 to confirm pluripotency and separated from mouse feeder cells using again a cell sorter, before storing them at −150°C21,22. The HUES9 hESC line (NIHhESC-09–0022) was also previously reported60, and single-cell-derived from sublines were generated using a cell sorter, as described above21,22. We have maintained their original names (HUES9 1.7, HUES9 1.8, and HUES9 1.9).
All hiPSC/hESC lines and sublines were cultured over a feeder layer of γ-irradiated MEFs, and maintained in hESC media made with knockout (KO) Dulbecco’s modified Eagle’s Medium (KO-DMEM) (Gibco, 10829–018) supplemented with 10% Plasmanate (Talecris Biotherapeutics); 10% Knockout serum replacement (KSR; Gibco, 10828–028); 20 mM GlutaMAX (Gibco; 35050061), 20 mM MEM nonessential amino acids (NEAA; Gibco, 11140050), β-mercaptoethanol (BME; Gibco 21985–023), and 20 mM Penicillin/Streptomycin (P/S; Gibco, 15140122); and 20 ng/μL fibroblast growth factor-basic (bFGF; Millipore; GF003AF-MG). Cells were passaged with Accutase (Innovative Cell Technologies, AT104) and maintained in hESC media supplemented with ROCK specific inhibitor Y27632 (Ri; Abcam-ab120129) for 24 hours, and then in hESC media with daily changes.
The mouse 46C mESC line (RRID:CVCL_Y482; breed: 129P2/Ola) and the ES-R1 mESC line (RRID:CVCL_2167; breed: 129X1/SvJ x 129S1/Sv-Oca2+Tyr+KitlSl-J) were previously reported61,62. Mouse ESCs lines were grown in feeder-free conditions, as previous described61. Mouse ESC media was prepared with DMEM-KO (Invitrogen 10829–018) supplemented with 15% ESC qualified-fetal bovine serum (FBS; Omega, FB-05) for mES-46C, or DMEM with high glucose and pyridoxine hydrochloride supplemented with 10% sodium pyruvate (Gibco 11360–070) and 20% FBS (Hyclone SH30070.03) for ES-R1. Both mESC media were supplemented with 1% NEAA (Gibco, 11140050), GlutaMAX (Gibco; 35050061), 1% P/S (Gibco, 15140122), 0.1 mM BME (Sigma, M7522), and 1μL/mL of recombinant mouse leukemia inhibitory factor (LIF; Millipore/Sigma, ESG1106) for ES-46C or 0.1 μl/mL of LIF (Millipore/Sigma, ESG1107) for ES-R1. Cells were grown in 0.1% gelatin (sigma) coated cell culture plates. To generate single-cell derived sublines, mES-R1 cells seeded on γ-irradiated MEFS were detached using 0.05% Trypsin (GiBCO, 25200–056) or Accutase (Innovative Cell Technologies, AT104). After centrifugation, cell pellets were resuspended in sorting buffer made of D-PBS containing 1mM EDTA (IBI Scientific, IB70184), 25mM HEPES pH 7 (Gibco, 15630–080), and 0.5 % BSA fraction V (Gibco; 15260–037). The cell suspension was filtered through tubes capped with filters (Falcon, 352235), and single cells were separated using a BD Influx Cell sorter and transferred to 96-well plates pre-seeded with γ-irradiated MEFS filled with 200μl of mES-R1 media supplemented with Ri (Abcam; ab120129) for 24 hours. Media without Ri was replaced daily for five-seven days. Colonies were detached with Accutase (Innovative Cell Technologies, AT104) to expand to larger wells in media supplemented with Ri (Abcam; ab120129) for 24 hours. The single-cell derived sublines generated from the parent EpiSC-R1 line were similarly established except that sorted single cells were seeded onto fibronectin-precoated 96-well plates and fed with mEpiSC media previously conditioned on γ-irradiated MEFS for two days and supplemented with 12 ng/mL bFGF (Millipore; GF003AF-MG), 20ng/mL Activin A (Peprotech; 120–14) before adding to cells. As above, when colonies were visible, they were passaged with Accutase (Innovative Cell Technologies, AT104) to expand, and fed with mEpiSC media supplemented with Ri (Abcam; ab120129) for 24 hours, and daily with only mEpiSC media afterwards.
Induction of 5iLA-naive identity
The induction of the 5iLA naive state in HUES9 and HUES9 1.8 cells was performed as previously described29,30 with a few modifications. Briefly, primed cells were seeded on γ-irradiated MEFS and maintained in hESC media for two days. Then, media was changed to modified primed media which is made as hES media but instead of 10% KSR (Gibco, 10828–028) and 10% plasmanate, contains 5% KSR and 15% FBS (Gibco: 16141079). After two-three days, cells were dissociated into single cells with Accutase (Innovative Cell Technologies, AT104) and passed through a 40 μm mesh-size cell strainer (Fisher Scientific, 22363547). Two hundred thousand cells were plated in one well of 6-well plate pre-seeded with γ-irradiated MEFS and maintained in modified primed media supplemented with Ri (Abcam; ab120129). Two days later, media was replaced with 5iLAF media that consisted of a 1:1 mixture of DMEM/F12 with GlutaMAX (Gibco; 35050061) and Neurobasal (Life Technologies), supplemented with 1x N2 (Gibco, 17502–048), 1x B27 (Gibco, 17504–044), 1x P/S (Gibco, 15140122), 1x NEAA (Gibco, 11140050), 0.5% KSR (Gibco, 10828–028), 0.1 mM BME (Gibco; 21985023), 50 μg/ml BSA Fraction V (Gibco; 15260–037), 20 ng/ml rhLIF (Preprotech; 300–05 or Millipore # LIF1005), 20 ng/ml Activin A (Peprotech; 120–14), 8 ng/ml bFGF (Millipore; GF003AF-MG), 1 μM MEK inhibitor PD0325901 (Stemgent), 0.5μM B-Raf inhibitor SB590885 (Tocris), 1 μM GSK3β inhibitor IM-12 (Enzo), 1 μM Src inhibitor WH-4–023 (Achemtek), and 10μM Ri (Abcam; ab120129). Eleven-twelve days post plating, cells were dissociated with Accutase (Innovative Cell Technologies, AT104) and re-plated on γ-irradiated pre-seeded plates after passing through a 40 μm cell strainer in 5iLAF medium. Naive hESCs were passaged with Accutase (Innovative Cell Technologies, AT104) cells every 5−7 days. After three-four passages domed-shape colonies were apparent. Naive cells were expanded to at least 3×10 cm plates to collect cell pellets for ChIP analysis or two 6-well for RNA-seq. Conversions were performed in a humidified 37°C incubator at 5% CO2 and under hypoxia (5% O2) or normoxia conditions (both conditions showed similar results). After 6–7 passages, naive HUES9 or HUES9 1.8 cells were transitioned to prime (re-primed) by switching 5iLA media to regular hES media and feeding lately with fresh media29,30.
Induction of EpiSC identity
For EpiSC generation, ES-R1 cells were passaged eight-ten times using Accutase (Innovative Cell Technologies, AT104), seeded in pre-coated dishes with 10–16 μg/mL human plasma fibronectin (Millipore-FC010) diluted in PBS, and fed daily with DMEM/F12 with L-glutaMAX (Gibco, 3050–061) base media supplemented with 0.5% N2 (Gibco, 17502–048), 1% B27 (Gibco, 17504–044), 50 μg/mL bovine serum albumin (BSA) Fraction V (Gibco, 15260–037), 1% NEAA (Gibco, 11140050), 1% P/S (Gibco, 15140122), 0.1 mM BME (Gibco, 21985–023), 12 ng/mL bFGF (Millipore; GF003-AF), 20 ng/mL Activin A (Peprotech; 120–14) as previously described63. Cells were passaged every two-three days, and Ri (Abcam; ab120129) was added for 24 hours after each passaging.
Generation of hiPSC-derived NPCs, neurons, or astrocytes
NPCs from CVB hiPSCs (parental line and single-cell derived hiPSC1–8 sublines) were generated using a protocol previously described21,22. Briefly, dissociated hiPSCs where plated at a very low density on a 10-cm dish pre-seeded with mouse PA6 stromal cells (5,000 hiPSCs/plate), and maintained in PA6 differentiation media made of Glasgow MEM (Gibco, 11710) supplemented 10% KSR (Gibco, 10828–028), 0.1 mM NEAA (Gibco, 11140050), 1 mM Sodium Pyruvate from Invitrogen, 0.1 mM BME (Gibco; 21985023), 0.5 mg/ml Noggin (R&D Systems), and 10 μM SB431542 (Tocris) for 6 days and then, fed every other day without SMAD inhibitors for another 6 days. Between twelve-fourteen days, neural rosettes were visible, and were dissociated with Accutase (Innovative Cell Technologies, AT104), and sorted (FACSARIA, BD Biosciences) based on a cell surface signature of CD184+/CD271−/CD44−/CD24+ using antibodies from BD Biosciences64. Sorted NPCs were plated on 20 μg/mL poly-L-ornithine and 5 μg/mL laminin (Life Technologies) coated plates in NPC medium containing DMEM/F12 with GlutaMAX (Gibco; 35050061) supplemented with 1% B27 (Gibco, 17504–044), 0.5% N2 (Gibco, 17502–048), 1% P/S (Gibco, 15140122), and 20 ng/mL bFGF (Millipore; GF003AF-MG). NPC media was changed every other day until confluency. NPCs derived from the hiPSC subline CVI 1.14 used in Supplementary Fig. 2b were similarly generated except neural rosettes were manually dissected21,22.
For neuronal differentiation, NPCs grown in 10-cm plates at confluency were switched to NPC medium without bFGF (day 1), and cultured for three weeks with one-two media changes a week, as previously described21,22. Around day 21, neurons were staining and sorted using the CD184−/CD44−/CD24+ signature64. Sorted neurons were plated on 20 μg/mL poly-L-ornithine and 5 μg/mL laminin pre-coated dishes in NPC media supplemented with 0.5 mM dibutyryl cyclic AMP (dcAMP; Sigma, D0267), and 20 ng/μL BDNF (Preprotech 450–02), and 20 ng/μL GDNF (Peprotech, 450–10).
For cortical differentiation, we followed a protocol previously reported65,66 with only minor modifications. Briefly, hiPSC1/3/5 were plated in a well of a 6-well plate pre-seeded with γ-irradiated MEFs and fed with hiPSC media supplemented with bFGF (Millipore; GF003AF-MG). At confluency, hiPSC were detached using Accutase (Innovative Cell Technologies, AT104) and transferred to a CELLstart CTS (Invitrogen A10142–01) pre-coated well of a 6-well plate and fed with hiPSC media supplemented with bFGF (Millipore; GF003AF-MG) and Ri (Abcam; ab120129). To initiate NPC differentiation, cell media was switched to NMM media made of 1:1 DMEM/F12:Neurobasal A supplemented with N2 (Gibco, 17502–048), B27 (Gibco, 17504–044), GlutaMAX (Gibco; 35050061), Insulin-Transferrin-Selenium-Sodium-Pyruvate (ITS-A; Gibco, 51300–044), P/S (Gibco, 15140122), BME (Gibco; 21985023), and NEAA (Gibco, 11140050), supplemented with 10 μM TGFβ-inhibitors SB431542 and 0.5 μM LDN-193189 (Stemgent; 04-001-05) for seven days changing media daily. Then, cells were detached with Versene and expanded to poly-L-Ornithine-Laminin coated plates in NMM media supplemented with SB and LDN. Next day, media was changed to NMM with no inhibitors and at the fourth day changed to NMM supplemented with bFGF (Millipore; GF003AF-MG) for two-three days before expanding again to poly-L-ornithine/Laminin pre-coated dishes, and fed with NMM with bFGF (Millipore; GF003AF-MG) for a few days. Neuronal rosettes should be visible at this point and NPCs can be frozen or continue to differentiate into neurons by bFGF withdrawal from NMM media.
For astrocytic differentiation, we also followed a protocol previously reported67. Briefly, NPCs derived from hiPSC1/4/5/8 were seeded in a 10-cm plate and grown in NPC media containing 20 ng/mL of bFGF (Millipore; GF003AF-MG) to confluency, scraped, and transferred to three uncoated wells of a 6-well plate. NPCs were grown in suspension under rotation (90 rpm) inside an incubator (37°C) and maintained with NPC media supplemented with 20 ng/mL of bFGF (Millipore; GF003AF-MG). The next day, when small floating neurospheres are observed, 5 μM Ri (Abcam; ab120129) was added to media for 24 hours. Two days later, the media was substituted to NPC media without bFGF (Millipore; GF003AF-MG) and changed every 2–3 days. One week after placing the cells on the shaker, the media was changed to Lonza Astrocyte Growth Media (AGM; Lonza) containing 3% FBS, ascorbic acid, recombinant human EGF, GA-1000 insulin, and L-glutamine (Lonza) maintaining the cells for an additional two weeks. Next, neurospheres contained in 3 wells of a 6-well plate were transferred to a poly-L-ornithine and laminin-pre-coated 10-cm plate. After one week (feeding cells every other day), astrocytes that have emerged from the neurospheres and are attached to the plate are dissociated with Accutase (Innovative Cell Technologies, AT104) and maintained in AGM media.
Generation of cortical organoids
For cortical organoid generation, CVB colonies were transitioned to feeder-free conditions as previously described34. Briefly, CVB colonies seeded on γ-irradiated MEFs were dissociated with Accutase (Innovative Cell Technologies, AT104), centrifuged at 1,000 rpm, and cell pellets were resuspended in hES medium (see above) pre-conditioned in γ-irradiated MEFs and supplemented with 20 ng/ml bFGF (Millipore; GF003AF-MG) right before seeding in pre-coated dishes with hESC-qualified matrigel (Corning) diluted in KO DMEM. Then, Ri (10 μM; Abcam; ab120129) was added for 24 hours. Colonies were grown daily and dissociated when confluent with Accutase (Innovative Cell Technologies, AT104) for at least three passages. Then, the media was substituted with mTeSR1 (STEMCELL Technologies) and cells were maintained in these conditions for at least two passages. From CVB colonies cultured and maintained in feeder-free conditions, we generated cortical organoids as reported34. CVB colonies maintained in mTSeR1 were dissociated with Accutase (Innovative Cell Technologies, AT104), centrifuged, and resuspended in mTeSR1 supplemented with 10 μM SB431543 (Stemgent), 1 μM Dorsomorphin (Tocris) and 5 μM Ri (Abcam; ab120129). Approximately, four-million cells were plated in a well of a six-well plate and grown in suspension under rotation (95 rpm) to form floating spheroids. Spheroids were fed daily with media without Ri (Abcam; ab120129) for two more days. Then, mTeSR1 media was substituted for neural induction media (Media1) containing Neurobasal (Invitrogen) supplemented with GlutaMAX (Gibco; 35050061), 15 mM HEPES, 1% NEAA (Gibco, 11140050), 1% P/S (Gibco, 15140122), 2% Gem21, 1% N2 (both STEMCELL Technologies), 10 μM SB431543 and 1 μM Dorsomorphin. Media1 was changed every other day for seven days, and then it was substituted for neuronal proliferation A media (Media2) made of NeurobasalA (Invitrogen) with GlutaMAX (Gibco; 35050061), 15 mM HEPES, 2% Gem21, 1% NEAA (Gibco, 11140050), 1% P/S (Gibco, 15140122), supplemented with 20 ng/mL bFGF (Millipore; GF003AF-MG) for seven days. Spheroids were split 1:2–3 and grown seven more days in Media2 supplemented with 20 ng/mL EGF (R&D systems) (neuronal proliferation B). Spheroids were split again, and neuronal maturation was initiated by substituting to Media2 supplemented with 10 ng/mL of BDNF, GDNF (both Peprotech), and NT-3 (R&D systems), 200 mM L-ascorbic acid (STEMCELL Technologies), and 1 mM dcAMP (Sigma, D0267). The media was changed every other day for six days; then, cortical organoids were switched to and maintained in Media2 for 10 months with media changes every 3 days. After day 56, only half of the media was changed every 3 days.
Grafting of hESC-Derived NPCs in rat spinal cords
We have complied with all relevant ethical regulations with regard to the use of animals in this study, which were approved by the University of California, San Diego Institutional Animal Care (Protocol: UCSD S01193). We generated NPCs for transplantation in rats from the ESI-017 (RRID: CVCL_B854) and H9 (RRID: CVCL_1240) hESC lines68,69. A thorough description of NPCs generation and procedures to graft hESC-derived NPCs into adult athymic RNU316 rats (Charles River, NIH) spinal cords has been recently described40. Briefly, hiPSC colonies were manually dissociated and induced to form embryonic bodies (EB). EBs were maintained in suspension with EB media. After four-six days, EBs were transferred to poly-L-polyornithine/Laminin pre-coated dishes and maintained in NSC media with bFGF (Millipore; GF003AF-MG) changing media every two-three days. Neuronal rosettes appearing 4–12 days after plating were manually dissociated and placed on poly-L-ornithine/Laminin coated dishes. This process of manual selection of rosettes was repeated twice to enrich for a population of enriched neuronal rosettes with small presence of cell of neuroectodermal origin. Then, a radially organized columnar epithelial cells outside the rosette structure were manually isolated and expanded to generate a highly homogenous and stable self-renewing population of NSCs. These NSCs were used for in vivo transplantation. Briefly, the day of surgery, ESI-017 or H9-derived NSCs were enzymatically detached from culture dishes, washed with and concentrated by centrifugation in hibernation buffer (HB; Neuralstem Inc., Rockville, MD), and stored on ice until grafting. Cells viability was measured prior surgery using Countess™ Automated Cell Counter (ThermoFisher). Rats were anesthetized, and submitted to partial laminectomy to expose the dorsal surface of L2–L6 segments as described70,71. Briefly, rats received ten to fifteen bilateral injections rostro-caudally every 700–900 μm of 0.5 μl containing either media alone or ~15,000 viable H9- or ESI-017-derived NSC using 80–100 μm in diameter glass capillary tips following established protocols70,71. The injection was targeted to deposit cells into the intermediate zone and the ventral horn of the spinal cord. Three weeks, two months or six months after grafting, rats were deeply anesthetized with pentobarbital and phenytoin and transcardially perfused with heparinized saline followed by 4% paraformaldehyde in PBS as previously described70,71. The spinal cords were dissected, postfixed, cryoprotected, cut on a cryostat, and stored in PBS with thimerosal at 4°C or frozen at −80°C for long time storage.
Immunohistochemistry of spinally grafted human cells
Extensive histological and immunohistochemistry characterization of grafted NPCs after different time points has been recently described40. Briefly, sections were stained with anti-human nuclear matrix protein/h-nuc (hNUMA; Millipore); anti-doublecortin (DCX; Santa Cruz), anti-human neuron specific enolase (hNES; Vector Laboratories), anti-human glial fibrillary acidic protein (GFAP; Origene), anti-choline acetyltransferase (ChAT; Millipore), anti-NeuN (Millipore), anti-human synaptophysin (hSYN; eBioscience), followed by fluorescently conjugated secondary antibodies as previously established procedures40,70. For long-time stored sections and to reduce autofluorescence sections were treated for one hour with sodium borohydride before staining. Images were captured using a confocal Fluoview F1000 or a Zeiss AxioImager M2 fluorescence microscope equipped with Apotome.2 for optical sectioning of images. Images were acquired and processed using Stereo Investigator software. Secondary antibodies: Cy3 Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch, 711-165-152), Alexa Fluor 488 Donkey anti-Mouse IgG (H+L) (Life Technologies, A21202), Alexa Fluor 647 Donkey anti-Rabbit IgG (H+L) (Life Technologies, A31573), Alexa Fluor® 594 Donkey Anti-Rat IgG (H+L) (Jackson ImmunoResearch, 712-585-153), Alexa Fluor 647 Donkey anti-Rabbit IgG (H+L) (Life Technologies, A31573), Cy2 Donkey Anti-Goat IgG (H+L) (Fisher Scientific, 705-225-147), Donkey anti-Goat IgG (H+L) Alexa Fluor 647 (Life Technologies, A21447), Alexa Fluor 647 Donkey anti-Chicken IgY (Fisher Scientific, AP194SA6MI).
RNA-seq and RT-qPCR analyses
For RNA-seq experiments, 1×106 CD184−/CD44−/CD24+ sorted neurons (see above) were plated on 20 μg/mL poly-L-ornithine and 5 μg/mL laminin coated 24-well plates and maintained in NPC media supplemented with 0.5 mM dAMP (Sigma, D0267), and 20 ng/μL BDNF and 20 ng/μL GDNF from Peprotech for 10–14 days. Total RNA was extracted using RNeasy Assay (Qiagen). RNA integrity number (RIN) was calculated using a TapeStation (Agilent Technologies) at the stem cell core of SCRM (UCSD). Most samples showed a RIN >7.0 and 7 out of 24 had a RIN between 6 and 7 (data not shown). PolyA+-RNA-seq libraries were generated using the Illumina TruSeq Stranded RNA LT kit and following manufacturer’s detailed instructions. A list of libraries can be found in Supplemental Table S2. Libraries were sequenced in an Illumina 2500 instrument at the UCSD IGM Genomics Center. Reads were aligned (one mismatch was allowed) to the hg18 human genome with bowtie2 (–very-sensitive option) and tophat (http://ccb.jhu.edu/software/tophat/index.shtml). Artifacts derived from clonal amplification were circumvented by considering maximal three tags from each unique genomic position as determined from the mapping data. Read counts were calculated with HOMER software (http://homer.salk.edu/homer/) considering only exonic regions of human RefSeq genes. Tags were normalized to 10 million reads per experiment. Normalization quality assessment was performed based on the subset of expressed RefSeq genes (≥50 counts) to ensure the assessment of the most informative gene subset. RNA-seq reads were visualized using the HOMER makeUCSCfile function. Counts per exon for RefSeq genes were calculated using HOMER. ‘Expressed RefSeq genes’ were defined as those (n=12,585) showing ≥50 counts, excluding n=11,168 RefSeq genes with <50 counts in the total of n=23,753 RefSeq genes (human genome, hg18).
For experiments in rats, all read alignments were performed with STAR. Counts to genomic regions were calculated using the Bedtools ‘coverage’ command using ‘-spilt and -abam’ options. For the rat/human split, raw FASTQ reads were first mapped to the Human genome (hg19). Successfully mapped reads were then mapped to the Rat genome (rn5). Reads that mapped to both genomes were evaluated by score and sorted to the genome to which they mapped with a higher score with a minimum score difference equivalent to 2 mismatches as well as a score exceeding some minimum score threshold for acceptance (equivalent to approximately 4 mismatches maximum). Reads that mapped to both genomes with similar scores were called ambiguous and are not included in either species split. Any reads that mapped to the Human genome but not to the Rat were sorted to Human. Finally, the reads that did not map to Human in the first step were mapped to Rat and all of those alignments exceeding the minimum score threshold are retained to Rat and others are called unaligned. The final Human and Rat sets of reads were combinations of those uniquely mapped to each species as well as those sorted to each species by score.
For RT-qPCR analysis, total RNA was also extracted using RNeasy Assay (Qiagen). DNA was removed using TURBO DNA-free kit (Ambion), and RNA was converted into cDNA using SuperScript First-Strand Synthesis System (Invitrogen) following manufacture’s protocols. PCR amplifications were carried out using the Applied Biosystems 7300 Real Time PCR System, FastStart Universal SYBR Green Master mix (Roche). ΔΔCt method was applied to obtain RNA expression levels, which were represented as fold change over a reference sample as indicated, or relative to a housekeeping gene as indicated. RT-qPCRs were performed at least three times (n≥3). Primers used for RT-qPCR analysis in this study can be provided upon request.
Single-cell (sc)RNA-seq analysis
For scRNA-seq analysis, we followed the protocol provided by Takara Bio USA Inc. in the SMART-seq HT Kit, which includes an adaptation of the NEXTERA XT DNA Library Prep protocol developed by Illumina. We isolated single neurons as described by the SMART-seq HT Kit protocol and processed 300 cells. We assessed the integrity of all the cDNA preps using the High Sensitivity DNA ChIP from Agilent Technologies, and only those samples with the bulk of signal between 600bp and 3,000bp were selected for further processing, which eliminated 104 samples. We processed 96 samples for library prep. We used two different DNA cleaning strategies: one strategy was based on pooling library preps before final cleaning (replicate 1); the second strategy was based on pooling library preps after final cleaning (replicate 2). Thus, only with the second strategy, we are sure that we sequenced the same amount of library prep for all samples. After sequencing of the two replicates of the same library for each cell (see RNA-seq, above), we obtained robust number of reads for 79 cells, but only for 74 cells in duplicate.
ChIP-seq analysis
For ChIP-seq, hiPSCs/hESCs seeded on γ-irradiated MEFs were grown to confluency, and mESCs were grown before reaching confluency. ChIPs were conducted as previously72,73. Briefly, cells were crosslinked for 45 minutes with 2 mM DSG (ProteoChem) diluted in 1xPBS buffer followed by 15 minutes treatment with 1% formaldehyde diluted in 1xPBS buffer (Sigma) for protein-based ChIPs. Both treatments were performed at room temperature under agitation, separated by three rounds of 1xPBS washes. Cells were crosslinked only with 1% formaldehyde for histone mark-based ChIPs. After crosslinking, cells were washed twice in 1xPBS and reactions were stopped with 100 mM Tris.HCl pH 9.4, 10 mM DTT. Fixed cells were harvested and frozen at −80°C for storage. For chromatin extraction, cells were lysed with lysis buffer (0.25% SDS, 10 mM EDTA, 50 mM Tris.HCl pH 7.8, inhibitor cocktail) at 4°C, and sonicated until chromatin fragments ranged lower than 1 kb size (in the absence of a decrosslinking step). Soluble chromatin was diluted 10x with dilution buffer (1% TritonX-100, 2 mM EDTA, 20 mM Tris.HCl pH 7.8, 150 mM NaCl, and protease inhibitors) and incubated overnight with antibodies, as indicated. The next day, 50 μL of 50% slurry of protein-A Sepharose beads (Sigma) were added and incubated for 6–8 hours, and finally washed twice with dilution buffer (15 minutes each time) and washed three times with 1xTE buffer. Decrosslinking was performed overnight at 65°C in 1% SDS, 1xTE buffer. DNA was purified with Qiagen purple columns. We used the following antibodies: H3K4me3 antibody (Millipore, 07–473), CTCF antibody (Millipore, 07–729), Rad21 antibody (Abcam, ab992), Sin3A antibody (Novus NB600–1263), Smch1 antibody (Abcam, ab31865), H3K9me3 antibody (Abcam, ab8898), Rest antibody (Millipore, CS200555), H3K4me2 antibody (Active Motif, 39141), and H3K9ac antibody (Millipore 07–352 and Abcam, ab4441), H2A.Z (Millipore, 07–594), and JMJD2A (Bethyl, A300–861A).
ChIP-seq libraries were prepared using the KAPA Library Preparation Kit (KK8201) as previously73,74. Samples were sequenced on Illumina 2500 and 4000 instruments and are listed in Supplemental Table S2. Single-end reads of 51 bp were generated. After mapping reads to the human genome (hg18) with bowtie2, artifacts derived from clonal amplification were filtered out by considering maximal three tags from each unique genomic position as determined from the mapping data. Peaks were identified with HOMER software (http://homer.salk.edu/homer/). Tags were normalized to 10 million reads per experiment.
Similarity Matrix Analysis (Heatmaps and Clustering)
Similarity matrixes (clustering and heatmaps) were generated using the Gene-E tool (version 3.0.204). Gene-E is a matrix visualization and analysis platform developed by Joshua Gould at The Broad Institute, available on-line at http://www.broadinstitute.org/cancer/software/GENE-E/. We used the Spearman rank correlation as metric for samples, or genes, or both, as indicated. Color scheme: relative, row minimum and row maximum, with selected values: −1 (dark blue, RGB: 0, 102, 204), −0.6 (light blue, RGB: 153, 204, 255), −0.2 (light grey, RGB: 245, 249, 241), 0.2 (light yellow, RGB: 255, 255, 153), 0.6 (orange, RGB: 255, 153, 0), and 1 (red, RGB: 255, 0, 0). We used RNA-seq samples, as indicated. We performed similarity matrix analyses with the list of expressed cPcdh isoforms, based on ≥30 counts in at least three preparations.
Radar plots of cPcdh expression
To generate radar plots, we only considered counts of the most 5’ variable exon of each isoform (Supplemental Table S3), excluding shared/common 3’ exons/UTR in the α/γ-clusters. For other genes, we considered exonic regions as indicated (Supplemental Table S3). Radar plots were generated in Excel (Microsoft) using normalized values to exon length (bp). cPcdh genes are listed (clockwise) in numerical and alphabetical order separating the three clusters.
Matching of RNA-seq and ChIP-seq Datasets (Violin Plots)
To match ChIP-seq and RNA-seq analyses, we organized variable cPcdh genes by groups based on levels of neuronal expression. For this analysis, we considered only tags derived from non-shared (5’) exons of the α/γ-clusters (Supplemental Table S3). Thresholds among groups were determined arbitrarily based on increasing counts: <30 reads, 30–100 reads, 100–600 reads, and >600 reads. RNA-seq values were averaged among triplicate preparations. In total, we established counts for n=384 genes/situations (n=48 c-PCDH genes in n=8 sublines). RNA-seq counts were matched with ChIP-seq counts corresponding to the corresponding stochastically selected promoters (Supplemental Table S4). In total, we established counts for n=384 promoter/situations (n=48 stochastically selected promoters in n=8 sublines), which were matched with the four groups based on increasing expression. Violin plots were generated using the VIOPLOT function in the violet package of the R software.
RNA-seq Meta-Analyses (Violin Plots)
Meta-analyses of c-PCDH expression were performed using normalized counts/bp of variable c-PCDH exons, housekeeping genes, fibroblast-specific genes, neuronal-specific genes, and pluripotency genes (Supplemental Table S3). List of previously reported datasets are indicated. Violin plots were generated using the VIOPLOT function in the violet package of the R software.
ChIP-seq Tag Density Meta-Profiles
Average tag density meta-profiles of ChIP-seq data were generated by counting sequencing tags in 10bp bins over 2kb, 4kb, or 6kb genomic windows, as indicated in the figures. Genomic coordinates were defined based on the distribution of all, H3K4me3, CTCF, and Rad21 ChIP-seq peaks, rather than based on the distribution of a single ChIP-seq peak (H3K4me3, CTCF, or Rad21), or the TSS annotation. Promoter meta-profiles were generated by averaging 2kb-wide ChIP-seq signal from the 13 alternate promoters in the α-cluster, the 16 alternate promoters in the β-cluster, and the 19 alternate promoters in the γ-cluster (Supplemental Table S4). These meta-profiles also included 25 2-kb-wide negative regions, in which peaks were not expected, as negative reference (Supplemental Table S4). For enhancers, profiles were generated form individual genomic regions (no meta-profiles). Data were smoothed by further averaging 5 adjacent bins (5 each side), and meta-profiles were generated with Excel (Microsoft).
DNA methylation analyses
Genome-wide DNA methylation data from two independent studies (GSE73747 and GSE63347) were reanalyzed44,45. GSE73747 was composed of Infinium 450K methylation data from 43 fetal frontal brains (16 Down syndrome and 27 control brains) and 11 adult brains (2 Down syndrome and 9 controls). Whereas, GSE63347 included Infinium 450K methylation data from 4 adult Down syndrome and 17 control frontal brains. Fetal Down syndrome and control brains: median=18-pcw/range=15–37-pcw and median=20-pcw/range=12–42-pcw, respectively. Adult Down syndrome and control brains: median=45 years/range=23–57 years and median=49.5 years/range=26–64 years, respectively. Array data were processed and normalized as detailed in the past44. Probes overlapping SNPs or located on sex chromosomes were excluded. In total, 465,572 probes met all quality criteria and had valid measurement in the two datasets. To account for potential batch effects, the ‘ComBat’ routine from the ‘sva’ package was applied after combining both datasets75. Subsequent methylation analyses focused on the promoter regions of the PCDH genes, defined as 2 kb upstream and 200 bp downstream from the TSS, leaving a total of 85 PCDHA, 105 PCDHB, and 151 PCDHG probes. Differential methylation estimates were obtained using a linear model based on methylation (beta) values averaged over each promoter as implemented in the limma package76. The model further included categorical parameters to adjust for batch and gender effects. All analyses were performed using the statistical software packages R (R version 3.2.2) in conjunction with the Bioconductor framework.
Statistical Analysis
We provide exact numbers for all statistics, unless the number of comparisons is too large to represent all P-values in the same figure (which is the case of Fig. 2b, in which we limit to show a range of significances; in this figure, in particular, we consider that it is not critical to show the exact number of the P-values, but to highlight the significant comparisons). RT-qPCR experiments were replicated in independent sublines and cultures, as indicated; in each case, technical replicates (n=3) were obtained. In bar plots, variation is indicated as mean ± standard error of the mean (s.e.m.) of technical replicates, and different bars represent different sublines and cultures. Sequencing experiments were performed in duplicate of independent cultures unless indicated. Violin plots show median, interquartile range (25th and 75th), and error bars represent 95% confidence intervals. In the DNA methylation analyses, the interquartile range (IQR) represents the 50% of the data, and the upper and lower quartiles represent the rest of the data (top 25% and bottom 25%, respectively). These analyses show mean differential methylation (of the indicated comparisons) with 95% confidence intervals (CI) on stochastically selected and non-stochastically selected α/γ-promoters. For immunostaining experiments, multiple images (n>3) were collected in each case and representative examples were shown. Statistical tests used: for correlation tests, we used the Pearson’s correlation analysis in the range (−1, 1), as indicated, with 0 indicating no association, and computed the associated P-value using cor.test function in R, testing the null hypothesis of the association being null; we conducted paired two-tailed Student’s t-tests or one-way ANOVA, multiple comparison test for statistical analyses using GraphPad Prism, as indicated. For the calculation of non-uniform versus uniform distribution of cPcdh isoforms in scRNA-seq, we used the Student t-test (t.test(A[2],alternative=”greater”): t=124.47; df=26457; alternative hypothesis (true mean is greater than 0); 95% interval: 35.969 (or any gene with occurrences >35.969 will be qualified as gene with significant high frequency); sample estimates: mean of x, 36.451. In this study, we did not make any particular assumption or remove data to perform our statistical analyses.
Reporting Summary
Further information on research design is available in the Nature Life Sciences Reporting Summary linked to this article.
Supplementary Material
ACKOWLEDGMENTS
We thank M.G. Rosenfeld and W. Dillmann for providing a nurtured research environment to perform this study, and the many colleagues who helped us to conduct or understand our research (in alphabetical order): A. Becskei, A. Gamliel, T. Haaf, A. Holder, K. Jepsen, E. Kothari, S. Linker, C. Marchetto, S. Marsala, J. Mertens, I. Narvaiza, F. Neri, D. Meluzzi, A. Muotri, P. Negraes, K. Ohgi, M. Parast, S. Sathe, D. Skowronska-Krawczyk, C. Trujillo, R. Tsunemoto, R. Van der Kant, G. Yeo, and the three anonymous reviewers. We also thank the team at the UCSD Human Embryonic Stem Cell Core Facility for reagents and technical assistance, and the ENCODE and BrainSpan Consortia for sharing valuable data. Special thanks to Honorio Garcia Garcia in representation of the many deceased anonymous donors who altruistically donate their bodies for the advancement of science. These donors have made possible our work. Q.M. was supported by the postdoctoral fellowship from the American Cancer Society. C.A. was supported by the postdoctoral fellowship Sara Borrell. Study supported by the U.S. Department of Defense (DoD) (AZ140064) to A.A.Q., the Sanford Stem Cell Clinical Center (SANPORC) to M.M., and NIH/NIA (1RF1AG048083-01 and 5P50AG005131-34) to L.S.B.G. The Department of Medicine, School of Medicine (UCSD) supported I.G.B.
Footnotes
DATA AVAILABILITY STATEMENT
The sequencing datasets generated in this study have been deposited in GEO under accession number GSE106872.
COMPETING INTERESTS STATEMENT
Martin Marsala is the scientific founder of Neurgain Technologies, Inc. and has an equity interest in the company. In addition, Martin Marsala serves as a consultant to Neurgain Technologies, Inc., and receives compensation for these services. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
REFERENCES
- 1.Gul IS, Hulpiau P, Saeys Y & van Roy F Evolution and diversity of cadherins and catenins. Exp. Cell Res. 358, 3–9 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hirayama T & Yagi T Regulation of clustered protocadherin genes in individual neurons. Semin. Cell Dev. Biol 69, 122–130 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Peek SL, Mah KM & Weiner JA Regulation of neural circuit formation by protocadherins. Cell. Mol. Life Sci 74, 4133–4157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mountoufaris G, Canzio D, Nwakeze CL, Chen WV & Maniatis T Writing, Reading, and Translating the Clustered Protocadherin Cell Surface Recognition Code for Neural Circuit Assembly. Annu. Rev. Cell Dev. Biol 34, 471–493 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Rubinstein R, Goodman KM, Maniatis T, Shapiro L & Honig B Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin. Cell Dev. Biol 69, 140–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett AM, Schreiner D, Lobas MA & Weiner JA γ-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron 74, 269–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostadinov D & Sanes JR Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T & Sanes JR Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molumby MJ, Keeler AB & Weiner JA Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity. Cell Rep 15, 1037–1050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mountoufaris G et al. Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356, 411–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Su H & Bradley A Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasic B et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Chen WV & Maniatis T Clustered protocadherins. Development 140, 3297–3302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y et al. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl. Acad. Sci. U.S.A. 109, 21081–21086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehayova P, Monahan K, Chen W & Maniatis T Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A 108, 17195–17200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monahan K et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of Protocadherin-α gene expression. Proc Natl Acad Sci U S A 109, 9125–9130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribich S, Tasic B & Maniatis T Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A 103, 19719–19724 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat. Genet 49, 1239–1250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Hajj N, Dittrich M & Haaf T Epigenetic dysregulation of protocadherins in human disease. Semin. Cell Dev. Biol 69, 172–182 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Woodruff G et al. Defective Transcytosis of APP and Lipoproteins in Human iPSC-Derived Neurons with Familial Alzheimer’s Disease Mutations. Cell Rep 17, 759–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff G et al. The Presenilin-1 ΔE9 Mutation Results in Reduced γ-Secretase Activity, but Not Total Loss of PS1 Function, in Isogenic Human Stem Cells. Cell Reports 5, 974–985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkelsen TS et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertens J et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell (2015). doi: 10.1016/j.stem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cacchiarelli D et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell 162, 412–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilens S et al. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat Commun 9, 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Leary T et al. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol 30, 278–282 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Warrier S et al. Transcriptional landscape changes during human embryonic stem cell derivation. Mol. Hum. Reprod (2018). doi: 10.1093/molehr/gay039 [DOI] [PubMed] [Google Scholar]
- 29.Theunissen TW et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 18, 262–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theunissen TW & Jaenisch R Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development 144, 4496–4509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yagi T Molecular codes for neuronal individuality and cell assembly in the brain. Front Mol Neurosci 5, 45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada T, Wallerich S & Becskei A Stochastic Gene Choice during Cellular Differentiation. Cell Rep 24, 3503–3512 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Trujillo CA et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell (2019). doi: 10.1016/j.stem.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JP et al. Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol 467, 1–10 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Stiles J & Jernigan TL The basics of brain development. Neuropsychol Rev 20, 327–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank M et al. Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell. Neurosci 29, 603–616 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Li Y et al. Synaptic and nonsynaptic localization of protocadherin-gammaC5 in the rat brain. J. Comp. Neurol 518, 3439–3463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou C, Huang W, Ying G & Wu Q Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience 144, 579–603 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Bohaciakova D et al. A scalable solution for isolating human multipotent clinical-grade neural stem cells from ES precursors. Stem Cell Res Ther 10, 83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyoda S et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82, 94–108 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhasova J et al. Time course of spinal doublecortin expression in developing rat and porcine spinal cord: implication in in vivo neural precursor grafting studies. Cell. Mol. Neurobiol 35, 57–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Hajj N et al. Epigenetic dysregulation in the developing Down syndrome cortex. Epigenetics 11, 563–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvath S et al. Accelerated epigenetic aging in Down syndrome. Aging Cell 14, 491–495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esumi S et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet 37, 171–176 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Smith A Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takashima Y et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borgel J et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet 42, 1093–1100 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Hasegawa S et al. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell. Neurosci 38, 66–79 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Hasegawa S et al. Distinct and Cooperative Functions for the Protocadherin-α, -β and -γ Clusters in Neuronal Survival and Axon Targeting. Front Mol Neurosci 9, 155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa S et al. Clustered Protocadherins Are Required for Building Functional Neural Circuits. Front Mol Neurosci 10, 114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katori S et al. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci 29, 9137–9147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad T & Weiner JA Direct and Indirect Regulation of Spinal Cord Ia Afferent Terminal Formation by the γ-Protocadherins. Front Mol Neurosci 4, 54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suo L, Lu H, Ying G, Capecchi MR & Wu Q Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol 4, 362–376 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Chen WV et al. Pcdhαc2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science 356, 406–411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamagishi T et al. Molecular diversity of clustered protocadherin-α required for sensory integration and short-term memory in mice. Sci Rep 8, 9616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gore A et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young JE et al. Elucidating molecular phenotypes caused by the SORL1 Alzheimer’s disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell 16, 373–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cowan CA et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med 350, 1353–1356 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Ying Q-L et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagy A, Rossant J, Nagy R, Abramow-Newerly W & Roder JC Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A 90, 8424–8428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo G et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan SH et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS ONE 6, e17540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi Y, Kirwan P & Livesey FJ Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 7, 1836–1846 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Shi Y, Kirwan P, Smith J, Robinson HPC & Livesey FJ Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci 15, 477–486, S1(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fong LK et al. Full-length amyloid precursor protein regulates lipoprotein metabolism and amyloid-β clearance in human astrocytes. J. Biol. Chem 293, 11341–11357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crook JM et al. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell 1, 490–494 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Mann DL et al. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res. Hum. Retroviruses 5, 253–255 (1989). [DOI] [PubMed] [Google Scholar]
- 70.Hefferan MP et al. Human neural stem cell replacement therapy for amyotrophic lateral sclerosis by spinal transplantation. PLoS ONE 7, e42614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kakinohana O et al. Region-specific cell grafting into cervical and lumbar spinal cord in rat: a qualitative and quantitative stereological study. Exp. Neurol 190, 122–132 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Almenar-Queralt A et al. Presenilins regulate neurotrypsin gene expression and neurotrypsin-dependent agrin cleavage via cyclic AMP response element-binding protein (CREB) modulation. J. Biol. Chem 288, 35222–35236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benner C et al. Decoding a signature-based model of transcription cofactor recruitment dictated by cardinal cis-regulatory elements in proximal promoter regions. PLoS Genet. 9, e1003906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.