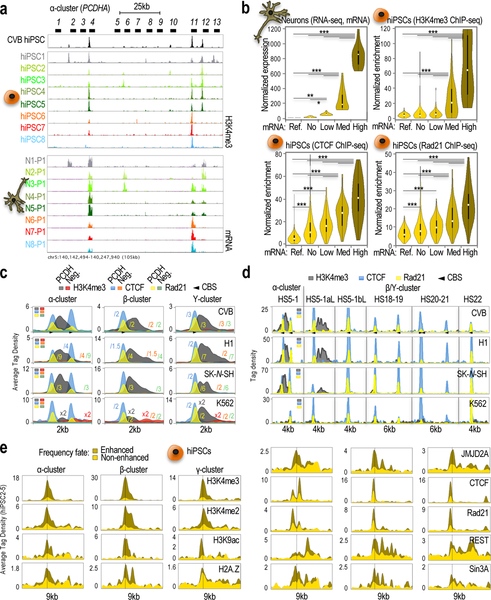
Fig. 2: Chromatin in hiPSCs mirrors expression in hiPSC-derived neurons.
a, ChIP-seq data showing H3K4me3 accumulation along the α-cPcdh cluster in the parent CVB hiPSC line and the n=8 different single cell-derived sublines (hiPSC1–8) matched with RNA-seq data showing α-cPcdh expression in n=8 hiPSC1–8-derived neurons (N1–8 P1; data from Fig. 1a). Scales=set to maximum (ChIP-seq) or identical (RNA-seq); 5’ α-cPcdh exons and genomic coordinates (hg18) indicated. b, Violin plots of matching RNA-seq signal (top-left; n=3 differentiation replicates for N1–8, n=24), H3K4me3 ChIP-seq signal (top-right, one culture from each subline; hiPSC1–8, n=8), CTCF ChIP-seq signal (bottom-left; two independent cultures from each subline; hiPSC1–8, n=16), and Rad21 ChIP-seq signal (bottom-right; two independent cultures from each subline; hiPSC1–8, n=16) stratified by groups based on neuronal expression levels (top-left). Groups: non-neuronal genes (n=11; “Ref”); “non”-expressed cPcdhs (n=193 out of 384, or 48×8); “low”-expressed cPcdhs (n=72); “medium”-expressed cPcdhs (n=57); and, “high”-expressed cPcdhs (n=26). One-way ANOVA, multiple comparison test, P-value<0.01 (*), <0.001 (**), or <0.0001 (***). Shown median, interquartile range (25th and 75th). Error bars represent 95% confidence intervals. c,d, 2/4/6kb-wide meta-profiles of H3K4me3/CTCF/Rad21 ChIP-seq averaged tag density along cPcdh promoters by cluster (in c), or at distal regulatory sites (in d) in the indicated cultures (experiments as in b). “Neg” represents random coordinates. Scale adjustments indicated (e.g. x2 and /2 refer to signal multiplied/divided by 2, respectively). CTCF sites/orientation indicated. e, 9-kb-wide meta-profiles of ChIP-seq data for activating and repressive components (n=4 ChIP experiments [hiPSC2–5] for component). Promoters segregated by H3K4me3 enrichment/non-enrichment (enhanced/non-enhanced, respectively).