Figure EV6. Differential methylation and copy number between healthy and tumor samples of tRNA genes, related to Fig 5 .
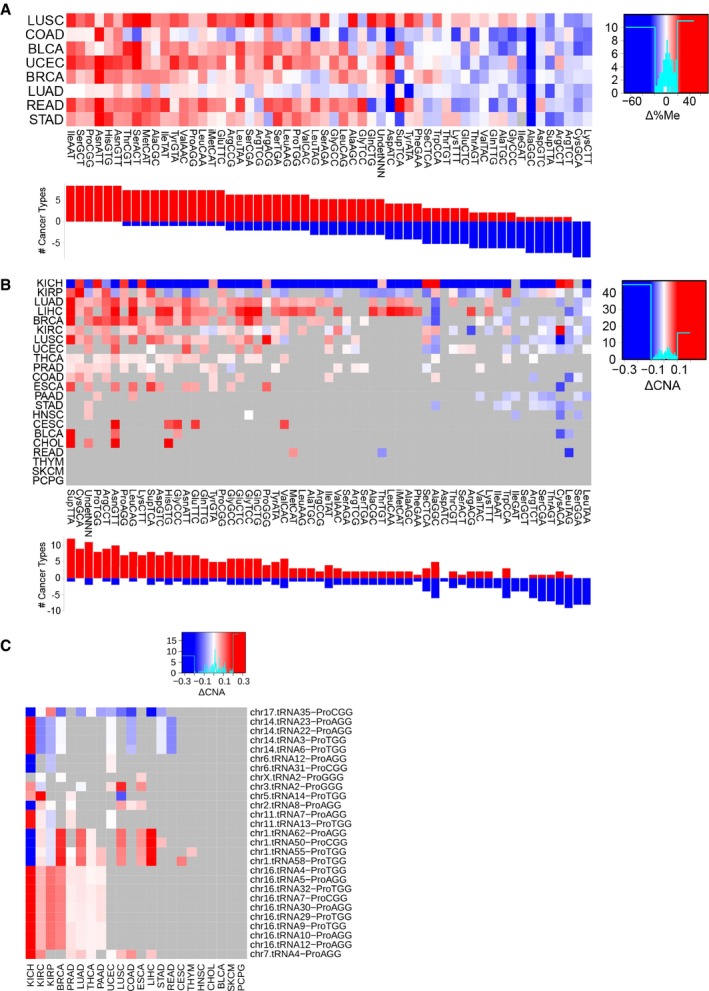
- Differential average methylation (bisulfite sequencing) for the isoacceptors, averaged over all genes that have a certain anticodon. The difference is measured by Δ%Me = (%MeTumor‐%MeHealthy). Refer to Table EV4 for full cancer type names and number of samples.
- Differential average gene copy number for the isoacceptors, averaged over all genes that have a certain anticodon. The difference is measured by ΔCNA = (CNATumor‐CNAHealthy). Only significant differences are colored, which are determined using a two‐tailed Wilcoxon rank‐sum test and corrected for multiple testing by FDR.
- Differential gene copy number between healthy and tumor samples of genes expressing proline tRNAs, as measured by ΔCNA. Only significant differences are colored, which are determined using a two‐tailed Wilcoxon rank‐sum test and corrected for multiple testing by FDR.
