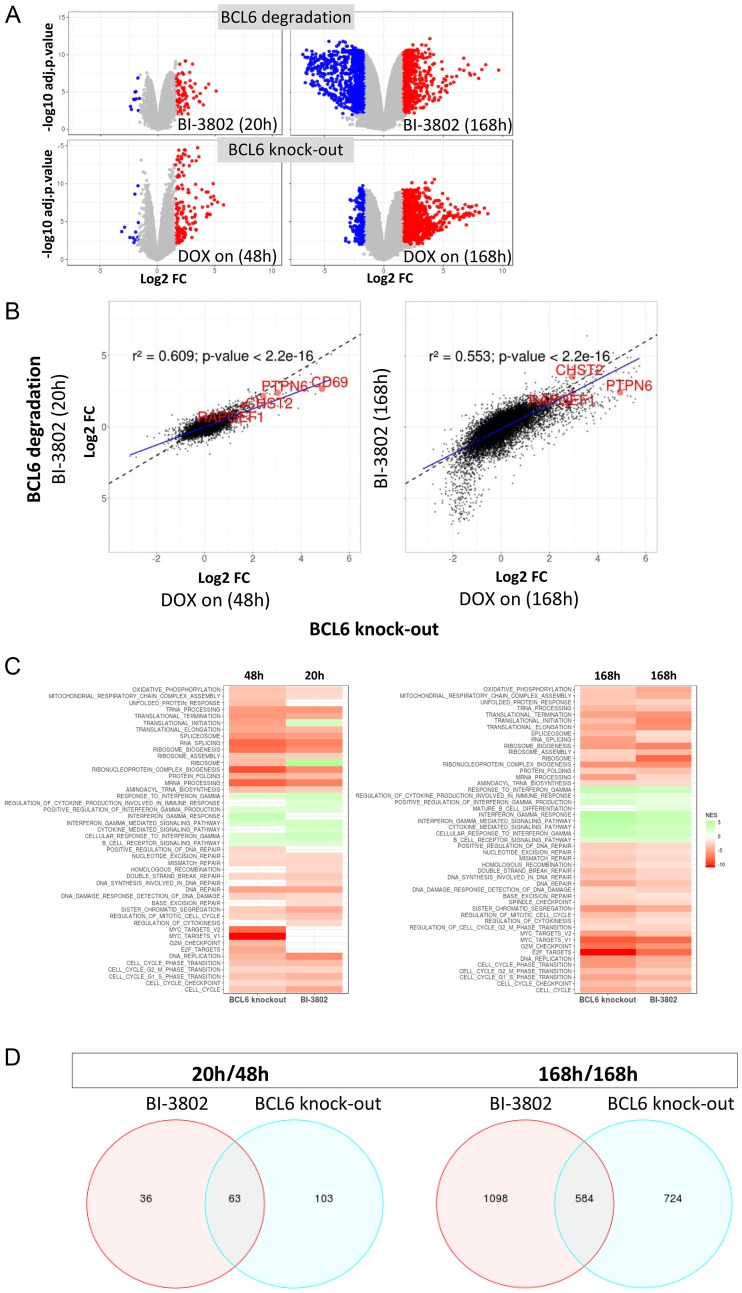
Figure 5. Conditional BCL6 knock-out in SU-DHL-4 induces gene perturbations similar to BCL6 degradation.
RNA-seq analysis was performed to compare gene expression after BCL6 knock-out and compound-induced degradation. (A) Volcano plot visualizing log2-scaled fold changes (x-axis) induced by either BI-3802 mediated degradation (compared to DMSO treatment) or BCL6 knock-out (compared to control sgRNA treatment) versus statistical significances (-log10 of the adj. p-value on the y-axis). Significantly deregulated genes (adj. p-value ≤ 0.01, fold change ≥ 3) are depicted in blue and red for repressed and induced genes, respectively. (B) Correlation of changes in gene expression induced by BCL6 knock-out (x-axis) or BI-3802 mediated degradation (y-axis). Genes near the dotted lines show comparable expression modulation in the BI-3802 treated data versus the BCL6 knock-out data set. Blue lines show linear regressions of the actual fold-change values. The goodness-of-fit of the linear regressions are shown by the r2 value in the graphs. (C) Gene set enrichment analysis (selected terms, FDR ≤ 0.1) reflecting genes set that are enriched/depleted for genes modulated by BCL6 knock-out or BI-3802 mediated degradation. The normalized enrichment score (NES) is color-coded in the heatmap. Negative values indicate gene sets that are significantly enriched for genes that are down-regulated upon BCL6 knock-out or BI-3802 treatment as shown in Supplementary Figure 4C (cell cycle). (D) Venn diagram indicating the overlap of genes after BCL6 degradation and BCL6 knock-out in SU-DHL-4 cells at the indicated time points of BI-3802 and DOX treatment.