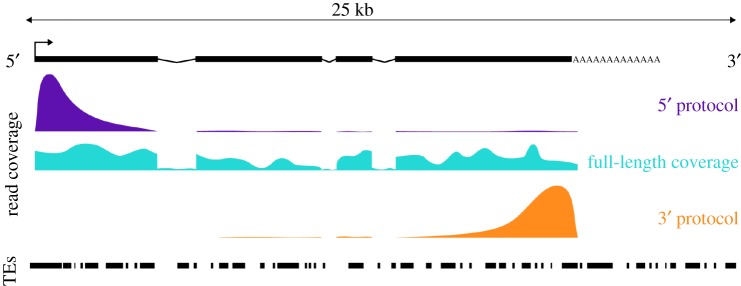
Figure 4.
Impact of different RNA-seq library strategies on read coverage along a TE-derived transcript with exon/intron structure (top). TE intervals are shown at the bottom. Briefly, scRNA-seq protocols that offer full-length transcript coverage provide the best means to identify full length transcribed TEs in a locus-specific manner, but this method suffers from noise owing to intronic TEs in host genes that might be mistaken for expressed TE transcripts as well as the inability to barcode individual mRNA molecules. 5′- and 3′-based protocols allow for barcodes that enable mRNA molecule counting, with 5′ protocols also offering the ability to detect TE transcripts originating from proper TE promoters.