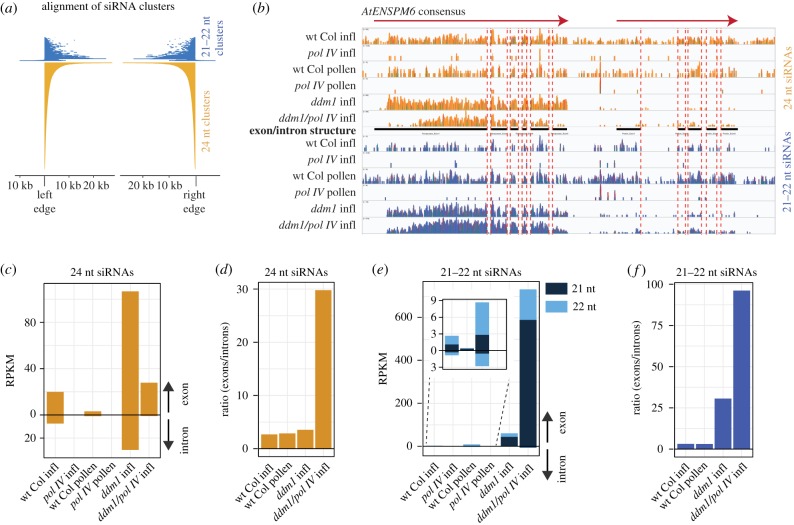
Figure 2.
Pol IV-dependent 21–22 nt siRNAs are produced from Pol IV transcripts. (a) Alignment of the 21–22 nt siRNA clusters to the ordered alignment of the 24 nt siRNA clusters. Very few 21–22 nt clusters extend beyond the boundaries of the 24 nt clusters, and inspection of these found no confirmable region of the genome that generates Pol IV-dependent 21–22 nt siRNAs but not 24 nt siRNAs in inflorescence tissue. (b) Alignment of siRNAs from various genotypes and tissues to the AtENSPM6 TE consensus sequence, which encodes two transcripts (red arrows). Black bars indicate protein-coding exons. Red dashed lines indicate transcript intron boundaries. Inflorescence is abbreviated as infl. (c) Reads per kilobase per million (RPKM) counts of intron- and exon-aligning 24 nt siRNAs for AtENSPM6. (d) Ratio of exon to intron small RNA abundance for 24 nt siRNAs. (e) RPKM counts of intron- and exon-aligning 21–22 nt siRNAs for AtENSPM6. Inset shows the low-level accumulation when TEs are not transcriptionally activated in the ddm1 mutant. Inset shows the low-level accumulation when TEs are not transcriptionally activated in the ddm1 mutant. (f) Ratio of exon to intron small RNA abundance for 21–22 nt siRNAs.