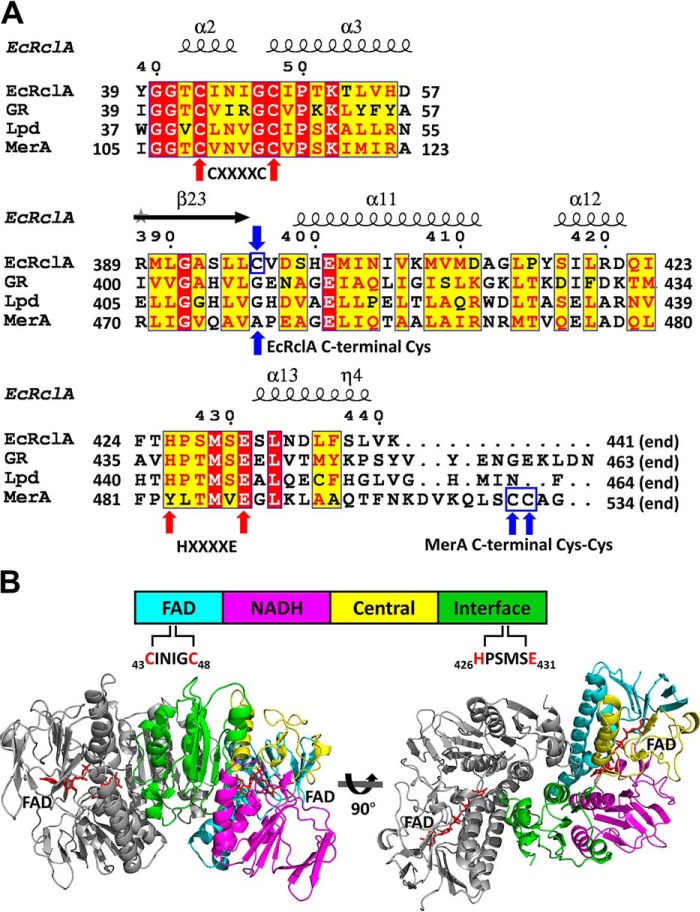
Figure 2.
Sequence analyses and the crystal structure of EcRclA. A, sequence alignment of FDR proteins with the secondary structure elements. GR, GSH-disulfide reductase from Bartonella henselae (group I FDR; GenBankTM accession no. WP_011180567); Lpd, dihydrolipoyl dehydrogenase from Mycobacterium multispecies (group I FDR; GenBankTM accession no. WP_003402301.1); MerA, mercuric reductase from Pseudomonas aeruginosa (group II FDR; GenBankTM accession no. WP_023980594.1). GR, Lpd, and MerA were chosen based on the highest score of DALI server structural analyses. The sequence alignment was performed using the T-Coffee server (35) and the ESPript server (36). For clarity, the regions containing the signature motifs are displayed. The full sequence alignment is presented in Fig. S2. The cysteine residues in the CXXXXC motifs and histidine and glutamate residues in the HXXXXE motif are indicated by red arrows. The C-terminal Cys in EcRclA and the C-terminal Cys–Cys motif in MerA are indicated by blue arrows and blue square. B, domain analysis and dimeric assembly of EcRclA. The schematic domain structure is shown at the top. The four domains are displayed in a bar containing a CXXXXC motif in the FAD-binding domain and the HXXXXE motif in the interface domain. The dimer is depicted by the ribbon representations at the bottom. One subunit is in the same color code of the top panel, and the other subunit is in gray. The bound FAD molecules are shown in the red stick representations.