Figure 3.
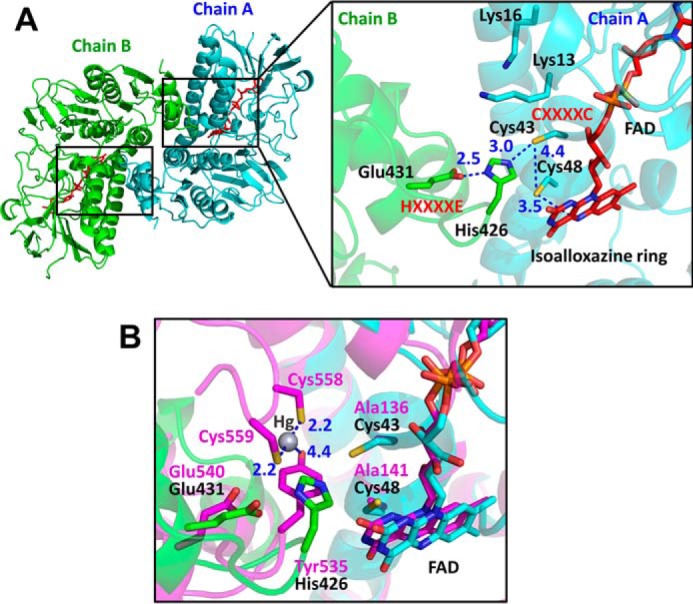
Structure of the active site of EcRclA. A, active site of RclA. Left, overall structure of RclA in the top view. Two identical active sites are marked as two small boxes. The box on the right is enlarged from one of the boxes from the overall structure on the left. Each subunit is depicted by the ribbon representation in cyan (chain A) or green (chain B). The bound FAD molecules are shown in the red stick representations. Each residue is labeled and indicated in the stick representations with the ribbon diagram in the background. The critical interactions between the two cysteines of CXXXXC from chain A, His-426 and Glu-431 of HXXXXE from chain B, and Cys-48 and the C4a atom of FAD are shown by blue dotted lines. Distances are in Å (blue). B, active-site comparison with MerA. The box is enlarged from Fig. S6. RclA (green and cyan) is aligned with MerA (magenta, PDB code 4K7Z) (37). Each residue aligned in the active site is labeled in a different color (black for RclA and magenta for MerA). His-426 in RclA is substituted with Tyr-535 in MerA. Mercury was bound to the two C-terminal cysteines of MerA, Cys5–58, and Cys-559. The interactions between the mercury, cysteine, and histidine residues are depicted by dashed lines. The two cysteine residues in the CXXXXC motif of MerA were substituted with alanine to determine the Hg2+-bound structure (37). Distances are in Å (blue).
