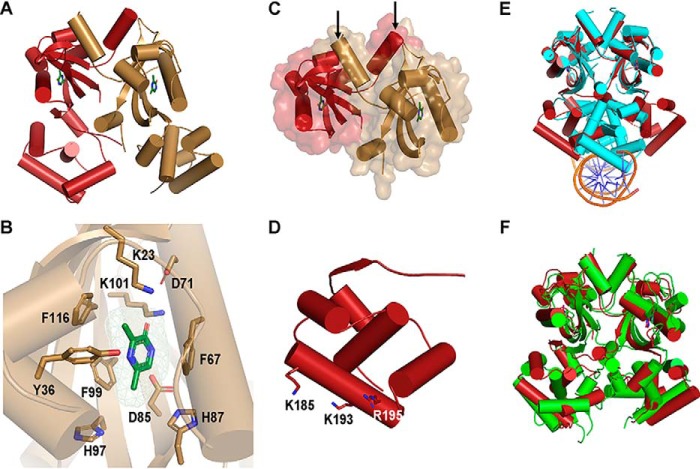
Figure 4.
Crystal structure of VqmA bound to DPO. A, 2.0 Å crystal structure of full-length VqmA as a dimer (monomers labeled in red and gold) with two molecules of DPO (green) bound. B, zoomed-in view of one monomer (gold) showing the DPO ligand (green) and its spatial relationship to the residues tested in the mutagenesis studies (Table 1 and Fig. S6). The DPO-VqmA interface is primarily stabilized by residues Phe-67, Phe-99, and Lys-101. C, the N-terminal PAS LBD of VqmA in cartoon format as in A, showing the buried surface area between monomers. The dimer is stabilized by the extensive buried surface region and crossover of two helices (highlighted by the arrows) at the N termini (residues 7–15) of the two monomers. D, the C-terminal HTH DBD of VqmA in cartoon format as in A. Based on solvent accessibility, Lys-185, Lys-193, and Arg-195 are proposed to be involved in DNA binding. E, structural comparison of DPO-VqmA (red) with a previously solved structure of DPO-VqmA bound to DNA (PDB code 6IDE (17); protein in cyan and DNA in orange and blue). F, comparison of the structure of DPO-VqmA solved here (red) with that of a recently reported structure (PDB code 6KJU (18); green).