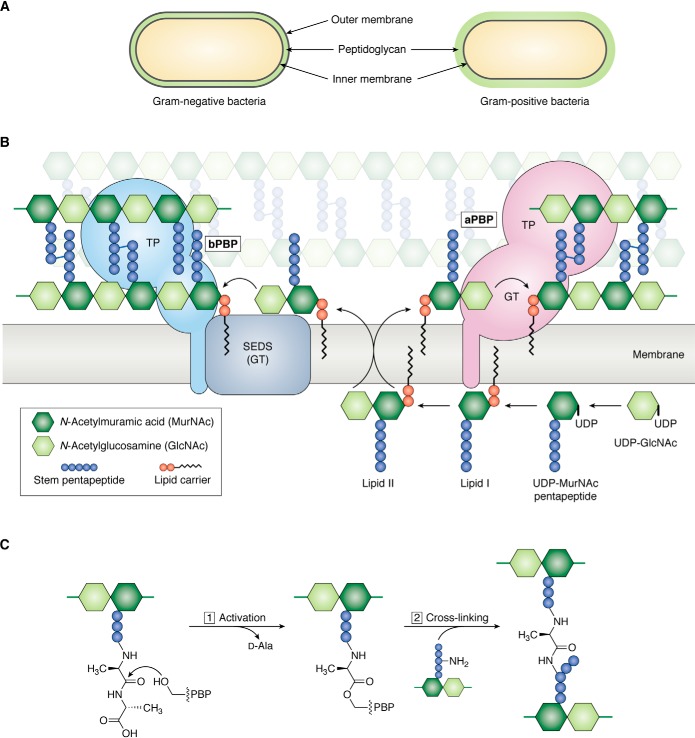
Figure 1.
Overview of peptidoglycan assembly pathway. A, Gram-negative bacteria have an inner (IM) and outer (OM) cell membrane; a thin layer of peptidoglycan is sandwiched between the cell membranes. Gram-positive bacteria only have an inner cell membrane that is surrounded by a thick layer of peptidoglycan. B, Lipid II, the undecaprenyl pyrophosphate lipid-linked precursor for peptidoglycan synthesis, is assembled on the inner leaflet of the inner cell membrane. Once fully assembled, Lipid II is flipped to the outer leaflet of the inner cell membrane by a flippase. Peptidoglycan GTs polymerize Lipid II into glycan strands, which are cross-linked by transpeptidases (TP) into the existing matrix. There are two families of glycosyltransferases: the GT module of aPBPs and SEDS proteins. Class A PBPs are bifunctional, meaning they have polymerization and cross-linking activities. SEDS proteins cooperate with a partner bPBP, which only has transpeptidase activity. C, the active-site serine of PBP transpeptidases attacks the terminal d-Ala–d-Ala amide bond in a stem peptide (donor), forming an acyl-enzyme covalent intermediate and kicking out the terminal d-Ala. Resolution of the covalent intermediate occurs upon reaction with a nucleophilic amine from an incoming stem peptide (acceptor), producing a new peptide bond that links two glycan strands.