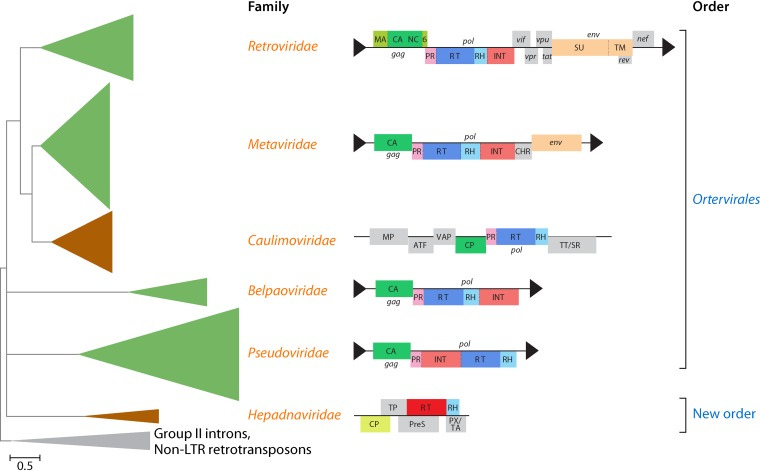
FIG 6.
Schematic phylogenetic subtree of the reverse transcriptases (RTs) of reverse-transcribing viruses (Caulimoviridae, Hepadnaviridae, and Retroviridae) and long terminal repeat (LTR) retrotransposons (Belpaoviridae, Metaviridae, and Pseudoviridae). The tree is rooted using sequences from nonviral retroelements (prokaryotic group II introns and non-LTR retrotransposons). Genome organizations of selected representatives of reverse-transcribing viruses and LTR retrotransposons are shown near the corresponding clades. LTRs are shown as black triangles. 6, 6-kDa protein; ATF, aphid transmission factor; CA/CP, capsid protein; CHR, chromodomain (contained only in the integrases of certain clades of metavirids from plants, fungi, and several vertebrates); gag, group-specific antigen; env, envelope gene; INT, integrase; MA, matrix protein; MP, movement protein; NC, nucleocapsid; nef, tat, rev, vif, vpr, and vpu, genes expressing regulatory proteins via spliced mRNAs; P, polymerase; pol, polymerase gene; PR, protease; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; RH, RNase H; SU, surface glycoprotein; TM, transmembrane glycoprotein; TP, terminal protein domain; TT/SR, translation trans-activator/suppressor of RNA interference; VAP, virion-associated protein. The two BCs included in the tree are color-coded (green, BC VI; brown, BC VII). (Adapted from reference 136.)