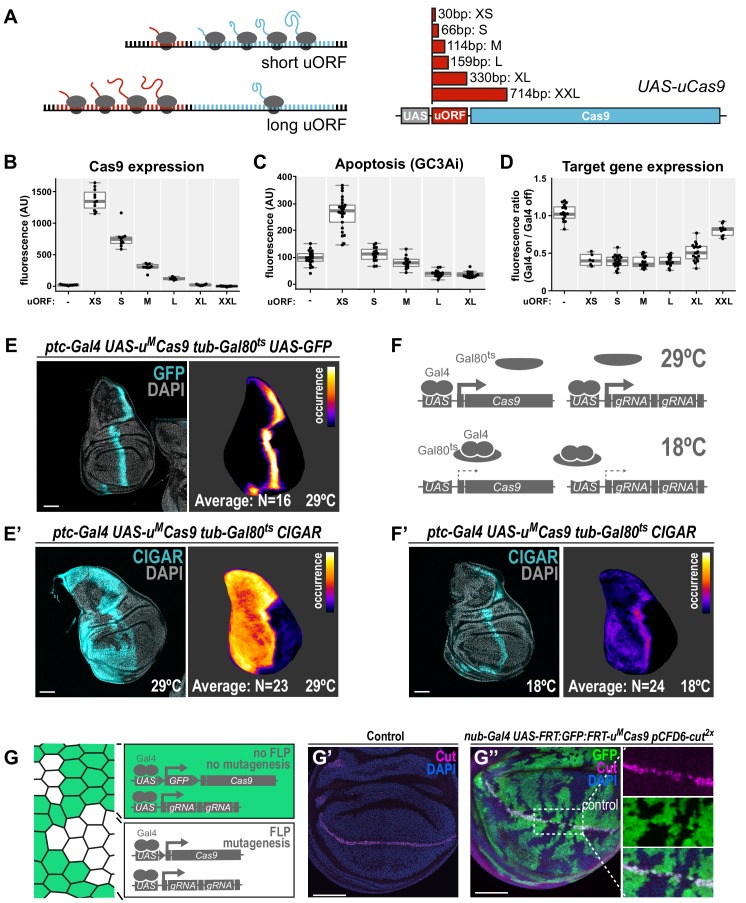
Figure 2. A transgenic series for tunable Cas9 expression to balance activity and toxicity.
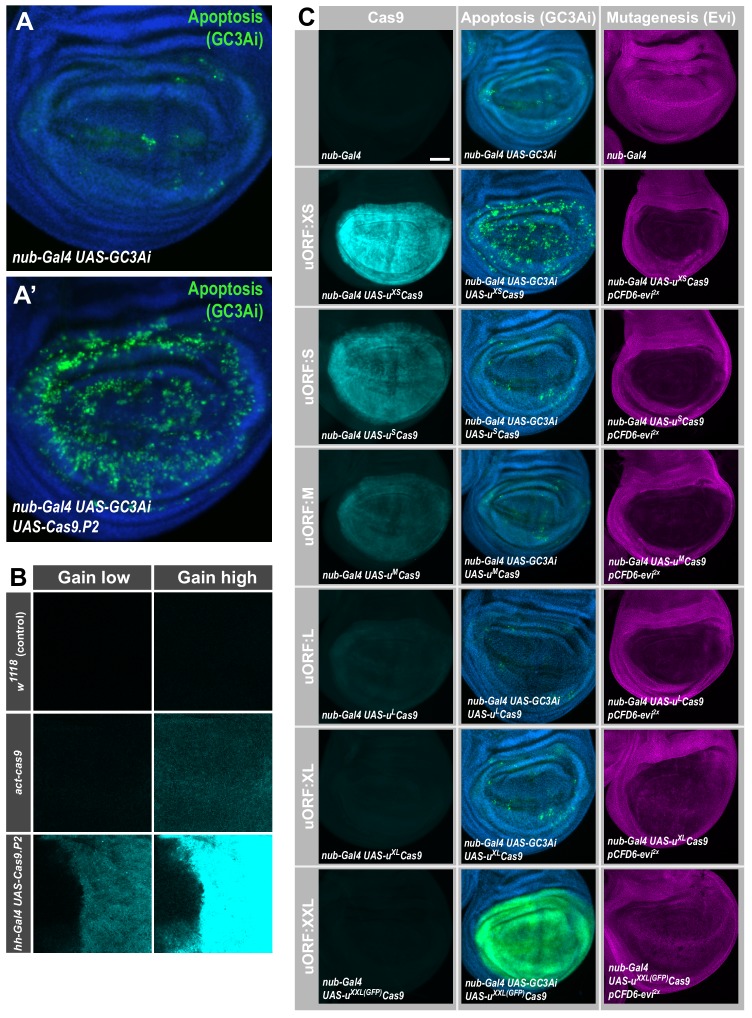
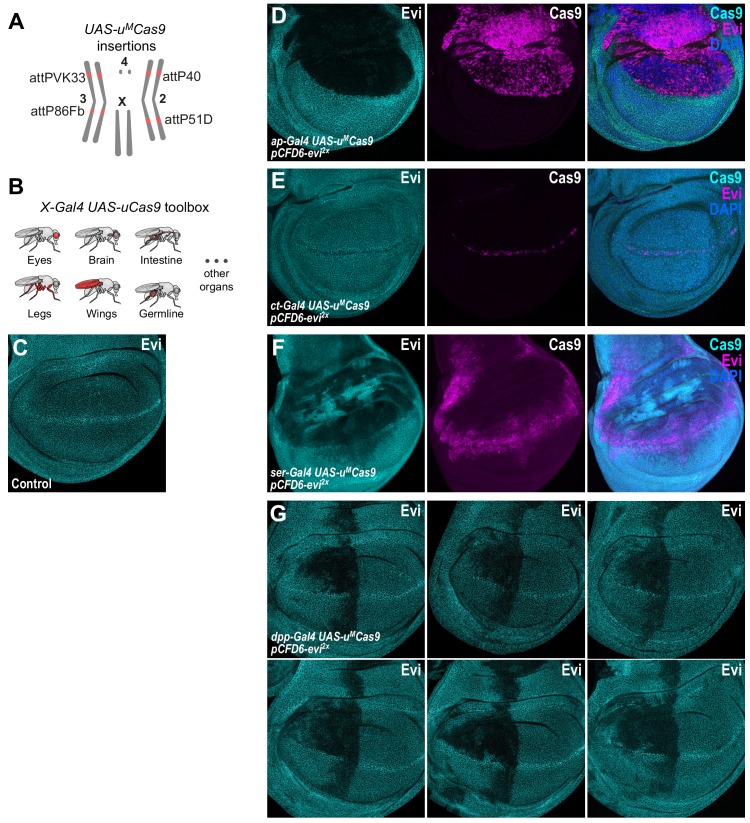
(A) Principle of the UAS-uCas9 series. Translation of the downstream ORF is inversely correlated with length of the upstream ORF in bicistronic mRNAs. The UAS-uCas9 series consists of transgenes that harbor uORFs of different length to modulate expression of Cas9. (B - D) Systematic characterization of Cas9 expression, toxicity and mutagenesis efficiency of the UAS-uCas9 series. Transgenes of the UAS-uCas9 series were recombined with nub-Gal4 and crossed to the apoptosis sensor UAS-GC3Ai (B, C) or pCFD6-evi2x (D). Graphs show data as individual dots, and boxplots as a data summary, with the line representing the median and the box the interquartile range. (B) Quantification of anti-Cas9 staining intensity in wing discs of the indicated genotype. Cas9 levels gradually reduce as the size of the uORF increases. N ≥ 6 wing discs. (C) Elevated levels of apoptosis were only observed with UAS-uXSCas9. The longest uORF (uXXL) encodes EGFP, preventing visualization of dying cells with GC3Ai. Quantification of fluorescent intensity of the GC3Ai reporter in the wing pouch. N ≥ 14 wing discs. (D) All transgenes of the UAS-uCas9 series mediate evi mutagenesis, with transgenes containing the four shortest uORFs (XS-L) leading to comparable gene editing that removes Evi from nearly all cells in the Gal4 expression domain. Quantification of staining intensity for Evi protein in the wing pouch (Gal4 on), relative to Evi staining in the hinge region (Gal4 off). N ≥ 6 wing discs. (E, E’) CRISPR mutagenesis patterns reflect Gal4 expression history. (E) Fluorescence of GFP, which turns over, reflects most recent Gal4 expression pattern. (E’) CRISPR mutagenesis, visualized by activation of the CIGAR reporter, is permanent and reveals the Gal4 expression history. Images of a representative wing disc are shown to the left of each panel and average intensity projection of several discs registered to a common template are shown on the right (see Materials and methods). Areas that are CIGAR positive in many discs appear bright, while areas devoid of signal in most discs appear dark. (F, F’) Incomplete repression of CRISPR mutagenesis by temperature-sensitive Gal80. (F) Principle of the Gal80ts system. At 18°C Gal80 binds and inhibits Gal4. (F’) Mutagenesis is still observed at 18°C in 11/24 discs and observed preferentially in the Gal4 expression domain, indicating incomplete Gal4 suppression by Gal80ts. (G - G’’) Control of CRISPR mutagenesis by a flip-out cassette. (G) In the absence of FLP recombinase a FRT-flanked GFP flip-out cassette (FRT sites represented by triangles) separates Cas9 from the promoter, resulting in cells that express GFP, but no Cas9. In the presence of FLP, the GFP cassette is excised and Cas9 is expressed. (G’) Staining for the transcription factor Cut reveals a continuous stripe of cells expressing ct along the dorsal-ventral boundary in wildtype tissue. (G’’) A pulse of FLP expression was used to excise the GFP flip-out cassette in a subset of cells (marked by the absence of GFP). Cut expression (magenta) is exclusively lost in GFP negative cells. Scale bar = 50 µm.