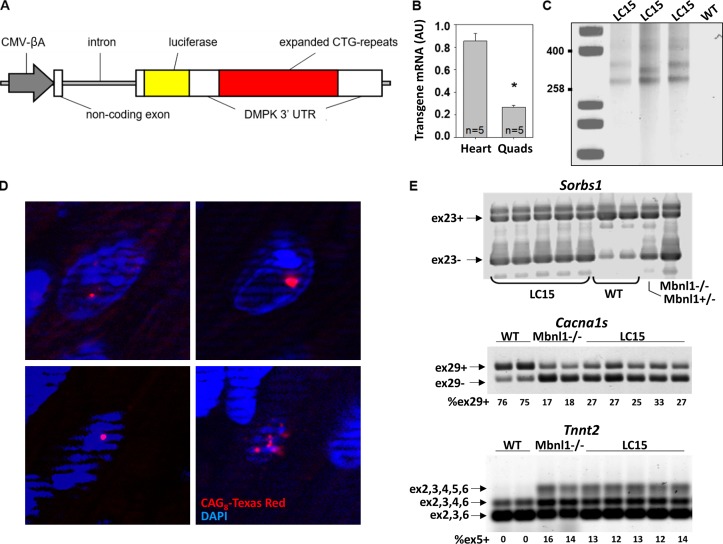
Figure 1.
LC15 transgenic mice exhibit robust CUGexp RNA expression in the heart. (A) Schematic of transgene used to generate LC15 mice. An uninterrupted expanded CTG-repeat tract generated using a cell-free assembly procedure described previously (Osborne and Thornton, 2008) was embedded in the natural context of the human Dmpk 3′-UTR. CMV-βA, CMV enhancer fused to chicken β-actin promoter. Luciferase is from Gaussia. (B) Summary of transgene mRNA expression by qRT-PCR (normalized to the housekeeping gene Gtf2b) in heart and quadriceps (Quads) muscle from five LC15 mice. The LC15 line displayed a higher transgene expression in heart (approximately fourfold) than skeletal muscle. (C) Southern blot of genomic DNA from three different LC15 mice (lanes 2–4) and one WT mouse (lane 5) used to determine CTG repeat size. Calibration controls (lane 1) indicate fragment size, expressed as number of CTG repeats. The CTG expansion ranges from 250 to 400 repeats. Note slight instability of the repeat tract in lane 3 compared with lanes 2 and 4. (D) FISH on four representative frozen sections of left ventricle from LC15 mice using a CAG8-Texas Red probe showing foci (red) of CUGexp RNA in nuclei (blue). Control section of WT muscle showed no signal (not shown). Nuclear foci are found in >80% of ventricular myocytes from LC15 mice. (E) Splicing of Sorbs1 exon 23 (top), Cacna1s exon 29 (middle), and Tnnt2 exon 5 (bottom) revert to their respective fetal patterns in hearts from five LC15 adult mice compared with that of two WT mice. Mbnl1−/− mice were used as controls for Mbnl1-dependent splicing.