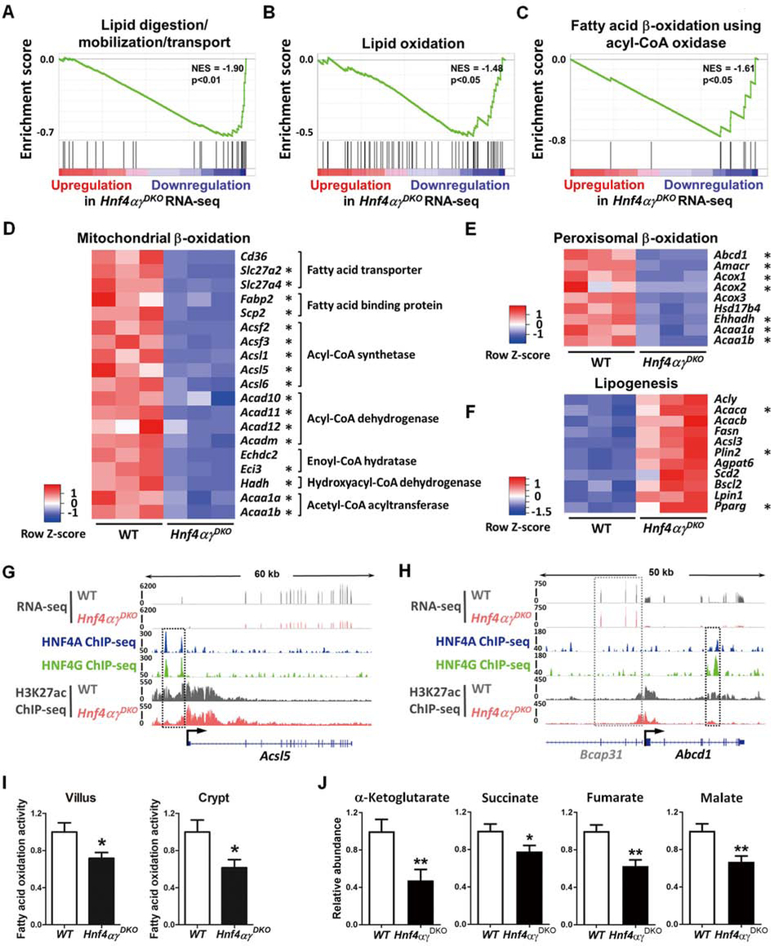
Figure 2. HNF4 paralogs bind to and activate FAO genes, and FAO is compromised upon HNF4 loss.
(A-C) GSEA of RNA-seq (duodenal epithelium, 2–3 days post tamoxifen-induced knockout, n=3 biological replicates, Kolmogorov-Smirnov test) reveals significantly reduced expression of genes related to lipid digestion and fatty acid β-oxidation in Hnf4αɣDKO. Heatmaps of RNA-seq data show (D-E) decreased transcript levels of mitochondrial and peroxisomal β-oxidation genes but (F) increased transcript levels of lipogenesis genes in Hnf4αɣDKO (n=3 biological replicates, FDR < 0.05). Asterisks indicate genes that are directly bound by HNF4 within 50kb of annotated TSSs by ChIP-seq. (G) ChIP-seq (n=2 biological replicates) and RNA-seq (n=3 biological replicates) tracks show that HNF4 factors bind to and activate Acsl5 (acyl-CoA synthetase gene), and loss of HNF4 results in reduced active chromatin signal (H3K27ac ChIP-seq, n=2 biological replicates, see black dashed rectangles). (H) HNF4 factors can also bind to and activate Abcd1 (peroxisomal β-oxidation related transporter). The neighboring gene locus is not bound by HNF4 and shows no change in expression levels, serving as an internal control (Bcap31, see gray dashed rectangles). More examples are shown in Fig. S2. (I) Hnf4αɣDKO mice show decreased FAO activity in both villus and crypt cells. The FAO assay measures palmitoyl-CoA-induced oxidation, as detected by NADH generation. Data are presented as mean ± SEM (villus, n=9 biological replicates; crypt, n= 3–5 biological replicates; Student’s t-test, two-sided at P < 0.05*). (J) TCA metabolites are reduced upon HNF4 loss. Metabolites were extracted from villus cells scraped from proximal small intestine of Hnf4αɣDKO and their WT littermates after 3 consecutive days of tamoxifen or vehicle injection, respectively. Corrected ion counts were normalized by tissue weight, and further normalized relative to the average abundance of metabolite from WT littermates of the same experimental batch. Data are presented as mean ± SEM (LC-MS, n=10 WT and 9 mutants, Student’s t-test, two-sided at P < 0.01** and P < 0.05*).