Abstract
Preclinical experiments and clinical trials demonstrated that Angiotensin II AT1 receptor overactivity associates with aging and cellular senescence, and that AT1 receptor blockers (ARBs) protect from age-related brain disorders. In a primary neuronal culture submitted to glutamate excitotoxicity, Gene Set Enrichment Analysis (GSEA) revealed expression of several hundred genes altered by glutamate and normalized by Candesartan correlated with changes in expression in Alzheimer’s patient’s hippocampus.
To further establish whether our data correlated with gene expression alterations associated with aging and senescence, we compared our global transcriptional data with additional published datasets, including alterations in gene expression in neocortex and cerebellum of old mice, human frontal cortex after age of 40, gene alterations in the Werner syndrome, rodent caloric restriction, Ras and oncogene-induced senescence in fibroblasts, and to tissues besides the brain such as muscle and kidney.
The most significant and enriched pathways associated with aging and senescence were positively correlated with alterations in gene expression in glutamate-injured neurons, and, conversely, negatively correlated when the injured neurons were treated with Candesartan. Our results involve multiple genes and pathways, including CAV1, CCND1, CDKN1A, CHEK1, ICAM1, IL-1B, IL-6, MAPK14, PTGS2, SERPINE1, TP53, encoding proteins associated with aging and senescence hallmarks, such as inflammation, oxidative stress, cell cycle and mitochondrial function alterations, insulin resistance, genomic instability including telomere shortening and DNA damage, and the senescent-associated secretory phenotype.
Our results demonstrate that AT1 receptor blockade ameliorates central mechanisms of aging and senescence. Using ARBs for prevention and treatment of age-related disorders has important translational value.
Keywords: Angiotensin receptor blockers, Glutamate excitotoxicity, p53 Neuroprotection, Aging, Senescence
1. INTRODUCTION
Aging is a universal process affecting the whole organism and leading to senescence, the loss of cell replication and enhanced apoptosis. Mayor processes compromising cells in the aging brain and leading to senescence include dysregulated inflammation, enhanced oxidative stress, activation of p53, cell cycle and mitochondrial dysfunction, insulin resistance, genomic instability with telomere shortening and DNA damage, cerebrovascular alterations and activation of the senescent-associated secretory phenotype [1–9]. In the brain, senescence impairs cognitive and motor skills and is the major risk factor for neurodegenerative disorders such as Parkinson disease and Alzheimer’s disease [10].
Angiotensin II, through AT1 receptor activation, contributes to homeostasis by regulating multiple brain functions [11–14]. However, brain AT1 receptor overactivity is associated with major alterations accelerating aging and leading to senescence, such as increased brain inflammation and oxidative damage, disruption of the mitochondrial respiratory chain, enhanced glutamate excitotoxicity and reduction of cerebral blood flow, major factors in the development and progression of age-related disorders [11–13, 15–17].
Consequently, inhibition of AT1 receptors by administration of Angiotensin Receptor Blockers (ARBs) has been shown to be strongly neuroprotective in neuronal, astrocyte, microglia and endothelial cerebrovascular cell cultures [18–21]. When administered in vivo, ARBs ameliorate early injury mechanisms including uncontrolled inflammation, age-related cognitive decline, prevent impairments in metabolic function and protect the cerebral vasculature [13, 17, 23, 23] Long-term inhibition of AT1 receptors by oral ARB administration in rats or life-long deletion of AT1 receptors in AT1 receptor knock out mice extend their lifespan [24–28].
ARB administration shares common protective and anti-aging mechanisms with calories restriction [20, 25] an intervention that extends the lifespan, reduces biomarkers of cellular senescence and retards several aspects of aging [29]. In addition, when administered systemically in vivo, ARBs ameliorate brain injury in rodent models of brain disease, particularly age-related cerebrovascular and neurodegenerative disorders such as Alzheimer’s disease and Parkinson’s disease [17, 19, 21, 27, 30–36]. These preclinical reports are being substantiated by controlled clinical studies, revealing that ARBs protect cognition after stroke and during aging, and by cohort analyses reporting that ARBs reduce the incidence and progression of Alzheimer’s disease [11, 12, 34, 37–40].
To address molecular mechanisms of ARB neuroprotection, we have performed a global gene analysis of rat primary neurons injured by glutamate [19]. The expression of hundreds of genes altered by glutamate was normalized when the neurons were incubated in the presence of Candesartan. When our findings were compared with those found in independently published datasets from Alzheimer’s disease autopsy brains [19], we discovered positive correlations in the expression of multiple genes altered by glutamate, and, conversely, negative correlations with the gene expression of glutamate-injured neurons treated with Candesartan. This study revealed common disease mechanisms between our in vitro results and those occurring in Alzheimer’s disease [19].
Since aging is the most important risk factor for Alzheimer’s disease, we asked the question whether Candesartan neuroprotection included additional pathways directly involved in aging and senescence. To this effect we run our global transcriptional data through Gene Set Enrichment Analysis (GSEA) in additional published datasets reflecting gene alterations during aging and senescence.
2. Methods
2.1. Gene expression analysis
We analyzed our own raw data from a previous experiment [19] submitted to Gene Expression Omnibus (GEO) under accession GSE67036. In this experiment we had incubated separated cultures of primary rat cerebellar granule cells (CGC) treated with either vehicle, Candesartan, glutamate or Candesartan and glutamate. Each group consisted on 5 independent experiments. Standard procedures extraction of total RNA, labeling, hybridization, washing and staining were as per manufacturer recommendation (Affymetrix, Santa Clara, CA). The raw data is submitted to GEO under accession GSE67036. Detailed procedures have been described [19].
2.2. Datasets description and microarray data mining
We used GSEA ( http://www.broadinstitute.org/gsea/) [41] to compare our data to published datasets [19]. For a more comprehensive description of the GSEA and the Broad Molecular Signatures Database v5.0 (MSigDB) [42] see [43, 44]. Alternatively, we used published microarray datasets from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) [45] to derive gene sets that we then used for GSEA analysis. All of the differentially expressed genes were included in the analysis: GSE8150 [46], GSE60652 [47], GSE28464 [48], GSE11882 [49], GSE17757 [50] and GSE11697 [51], were downloaded from the National Center for Biotechnology Information (NCBI) GEO database and imported into Partek Genomics Suite software (Partek, Inc., St. Louis, MI). The geneset names, accession numbers and website links are all described in Table 1 and Online Resources; 1 for aging, 2 for senescence and 3 for combined brain regions). Ingenuity pathway analisis (IPA). (http://www.ingenuity.com) [52] (Ingenuity Systems, Redwood City, CA) was used to identify canonical pathways associated with the differentially expressed genes.
Table 1.
Genesets names, accession IDs, and links for the 19 genesets analyzed in this manuscript.
To identify pathways associated with differentially expressed genes we used the CluePedia plugin of Cytoscape (v3.7.1) (http://www.cytoscape.org/) [53]. Datasets downloaded from GEO were Robust Multichip Average (RMA) normalized and differential gene expression was accessed by one-way ANalysis Of VAriance (ANOVA). For microarrays cross platform comparisons, we used a p value of <0.05 and a 1.2-fold change cutoff. Raw data from these datasets were analyzed with Partek Genomics Suites under similar conditions used for our neuronal culture data. To avoid cross platform heterogeneity, we focused only on datasets generated on the Affymetrix platform.
3. RESULTS AND DISCUSSION
We run our global transcriptional data (GSE67036) [19] through the GSEA (http://www.broadinstitute.org/gsea/ [42]. Using this functional enrichment analysis, we identified multiple genes altered in our study that were over-represented and most significantly associated with alterations in gene expression found in many aging and senescence datasets. Multiple genes in these selected genesets, that were associated and upregulated with aging and senescence (Fig 1 and Online Resources 1 and 2) showed a statistically significant positive correlation with genes upregulated by glutamate in our study and a negative correlation with the expression of those genes that, when upregulated by glutamate, were reversed or normalized by Candesartan. Based on these associations, we postulate that administration of Candesartan could significantly ameliorate or reverse the effects of aging and senescence in gene expression.
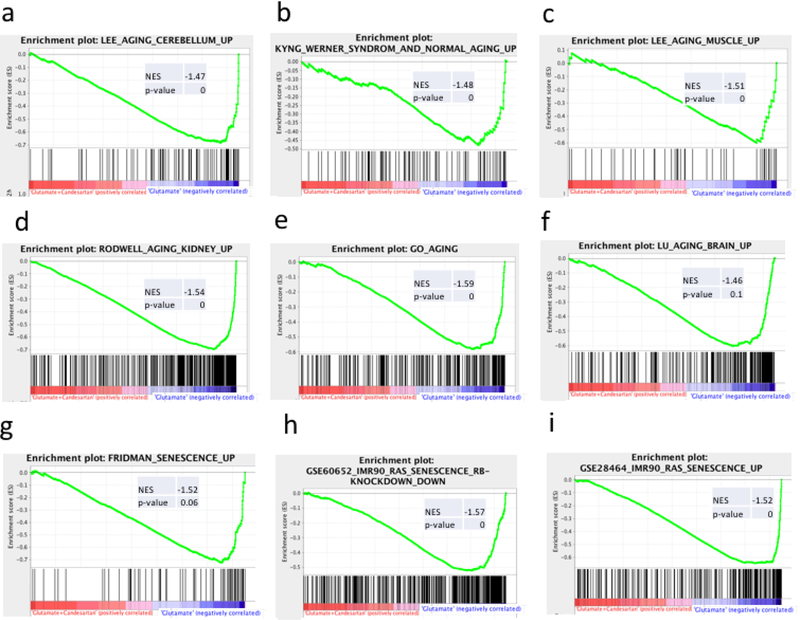
Fig 1. Gene Set Enrichment plots from representative aging pathway genesets.
The Fig represents Gene Set Enrichment Analysis plots showing the negative correlations of genes up-regulated by Glutamate and normalized by Candesartan in our neuronal cell culture with genesets associated with general aging in different brain areas and tissues (A-F, see Online Resource 1) and with senescence (G-I, see Online Resource 2). NES: Normalized Enrichment Score. Genesets with links and References are listed in this Table1.
The results obtained with our cell culture were compared with many transcriptome analysis studies in mice. Analysis of gene expression profiles of neocortex and cerebellum in adult and old mice indicated alterations in the expression of multiple genes with aging, that were similar in these two regions and that were indicative of inflammatory responses, microglial activation, complement cascade genes, stress response, oxidative stress, and reduced neurotrophic support [54] (Fig 1a; Online Resource 1). The association with changes in the expression of genes in our study were similar in all cases. There were 38 out of 85 enriched genes when comparing those found upregulated in the aged neocortex including NDRG1, FOS, CRYAB, APOD, MPEG1, C1QC, IFI27, C1QA, PTGS1, CTSZ, CTSS, (Online Resource 1), and 26 out of 80 genes upregulated in the aged cerebellum (LEE_AGING_CEREBELLUM_UP) including FOS, IRF7, MPEG1, C1QC, BCL2A1, IFI27, CIQA, AXL, CTSZ, and CTSS. (Fig 1a, Online Resource 1). This study also noted that alterations in gene expression in brains from aged mice correlated with those found in autopsy brains from patients who suffered from neurodegenerative disorders [54].
A comparison of the gene expression profiling of fibroblast cell lines from young and old human donors with those obtained from patients suffering from Werner Syndrome or progeria, a premature aging disorder, indicated that over 90% of the annotated genes were similarly changed in normal aging [55]. Our data showed a striking correlation with 19 genes out of 83 altered in both Werner syndrome and normal aging. Genes up regulated in Werner Syndrome and normal aging are also upregulated by glutamate and normalized by Candesartan in our neuronal culture, including CHEK1, NDRG1, TNFRSF10B and NR1H3. (Fig 1b, Online Resource 1; KYNG_WERNER_SYNDROM).
These results are supported by a meta-analysis of multiple age-related gene expression profiles consistently identified with age [56]. In this study, 29 out of 49 identified genes were enriched in our microarray analysis including LITAF, APOD, MPEG1, C1QC, C1QA. CTSS, CTSZ (Online Resource 1). Another study analyzed the transcriptional profile from the neocortex of young, adult and old mice treated with vitamin E supplementation [46]. In this study, 122 out of 422 genes upregulated in the cortex of aged mice correlated with our microarray analysis including GFAP, ZCCHC24, EFS, HSPA2, LITAF, CCND1, C1QA, IFI27, C1QC, GRN, APOD and MPEG1 (Online Resource 1).
The correlation of our neuronal culture results and those revealed in aging is not limited to findings in the brain, but they are also consistent with gene alterations reported in other tissues such as muscle including TGFB1I1, FOS and TGIF1 [57] (Fig 1c, Online Resource 1), and NRG1, FGFR2, TFAP2A, CNP, GRN, CCL2, CDKN1A, PIK3CG and SERPINE1 [58] Online Resource 1, and kidney, including RAB31, ZCCHC24, TIMP2, MPEG1, GRN, C1QC, MYOF, BCL2A1, AXL, C1QA, CCL2, VCAM1 and SLC6A6 [59] (Fig 1d, Online Resource 1).
Similarly, two analysis of genes upregulated in general cellular aging (From the Gene Ontology (GO) database http://www.ncbi.nlm.nih.gov/geo/) [45] revealed that 90 of 252 genes, (Fig 1e, GO_AGING, Online Resource 1) and 22 out of 62 genes are enriched when compared with our microarray analysis (GO_CELL_AGING. Online Resource 1), respectively. This includes TP53, HMGA1, CHEK1, LITAF, FOS, TIMP2, CRYAB, MME, TNFRSF10B, APOD, SERPINE1, IL6 and IL1β.
A transcriptional profile in human frontal cortex was compared in human autopsy samples ranging from ages from 26 to 106 years. This study reported that the expression of a set of genes important for synaptic plasticity, vesicular transport, mitochondrial function, calcium homeostasis and neuronal signaling and involved in learning, memory and neuronal survival was reduced, and that DNA damage was enhanced, with reduced DNA repair in their promoters after age 40. These findings correlated with those damaged by oxidative stress in cultured human neuroblastoma cells and are indicative of a genetic signature of human cortical aging [60]. A comparison of these results with our global gene analysis revealed that of the 244 genes altered in the aged human frontal cortex, the expression of 129 genes negatively correlated with our findings in neuronal cultures [60] (Fig 1f, LU_AGING_BRAIN_UP, Online Resource 1).
Genes reduced in the aged cortex and damaged by oxidative stress reduce DNA repair, leading to decreased expression of genes involved in learning, memory and neuronal survival. These include genes such as NDRG1, GSN, C1QC and C1QA, involved in defects of the DNA-repair system, the tumor suppressor pathway, the telomere maintenance system, the insulin/Akt pathway, and other metabolic pathways [60].
Of special interest is that alterations in gene expression in our neuronal culture dataset affected by glutamate and normalized by Candesartan strongly associate with those found in two mice strains subjected to calorie restriction [57]. It is already known that enhanced calorie intake is a risk factor for diabetes and hyperinsulinemia, and that alterations of the insulin pathway induce cellular senescence in vitro [61]. In turn, calorie restriction, like long-term ARB administration, extends the lifespan of various species, favors lipid metabolism, protects from renal disease, decreases biomarkers of cellular senescence in vivo, and retards several aspects of aging [15–17,29]. Calorie restriction reduced the age-associated up-regulation of genes regulating inflammation, stress, microglial activation and the complement cascade [54]. We have recently shown that changes in gene expression during calorie restriction associate with the expression of genes altered by inflammation in microglia cultures and normalized by the ARB Telmisartan [20]. The effects of calorie restriction are not restricted to the brain, since it reduces age-dependent oxidative damage in muscle [57, 58] Online Resource 1). These results support the hypothesis that ARB administration and calories restriction share common protective and anti-aging mechanisms, and they are both associated with longevity.
The correlation of our neuronal culture results is not limited to published aging-related databases but extend to databases reporting alterations in gene expression involved in senescence. A review of studies on gene expression profiling to identify those genes involved in senescence, some with supporting functional data, revealed universal genes regulating senescence, across cell types and model systems, including the cell cycle pRB/p53, cytoskeletal, interferon-related, insulin growth factor-related, MAP kinase and oxidative stress pathways [62], In this dataset, of the 75 genes upregulated by senescence, 37 genes up regulated by glutamate and down regulated by Candesartan were found enriched in our glutamate-candesartan study (Fig 1g, Online Resource 2). They include STAT1, SERPINE1, RAB31, TFAP2A, TGFB1/1, HSPA2, IRF7, CRYAB, CTGF, GSN, CDKN2B, THBS1, CYP1B1, THBS1 and CDKN1A.
We found additional correlations between our neuronal culture results and a microarray analysis of human IMR90 fibroblasts differentiated into macrophages submitted to Ras-induced senescence [48]. From the many genes (378) with enhanced expression by senescence, the expression of 135 genes was positively correlated in our study with glutamate-induced injury and negatively correlated to the effects of Candesartan in our system (Fig 1h), including TGFB1/1, HSPA2, LPP, CTGF, MYOF, THBS1 and PTGS1 (Online Resource 2 GSE60652_IMR90_RAS_SENESCENCE_R)
Similarly, microarray analysis of IMR90 fibroblasts subjected to oncogene-induced senescence with reduced glycolytic pathway and down regulation of the retinoblastoma protein identified 82 genes out of 254 are enriched in our neuronal cultured study [47] (Fig 1i). (Online Resource 2 GSE28464_IMR90CELLS_RAS_SENESCENCE_UP)
Gene Ontology analysis of genes associated with senescence identified that out of 30 senescence genes, 18 genes were enriched in our study (GO_CELLULAR_SENESCENCE, Online Resource 2). Genes upregulated by glutamate, normalized by Candesartan and enriched in this study include CDKN1A, TP53, MAPK14, CAV1, THBS1 and HMGA2.
Moreover, using proliferating and senescent WI-38 cells, [63] identified 31 and of those 8 were enriched in our glutamate-Candesartan analysis, including CDKN1A that encodes for p21, a major target of p53 and linking DNA damage to cycle arrest [64], LCAT, CCND2, stimulated by oxidative stress inducing cellular senescence [65] and ADAMTS5, a disintegrin and metalloproteinase with thrombospondin motifs 5, upregulated in extracellular matrix destruction and osteoarthritis [66] (Online Resource 2, TANG_SENESCENCE_TP53_TARGETS_UP).
Given the large number of aging genesets negative correlated with glutamate + Candesartan, we sought to identify common aging genesets so we could define pathways associated with Candesartan effects. We used the GEO database to compare our dataset with up-regulated genes in 6 different published aging datasets from different human and primate brain areas. GSE11882 included samples from the human posterior cingulate cortex (PCG), superior frontal gyrus (SFG), Hippocampus (Hippo), and Entorhinal Cortex (IEC) [49]. GSE11697 included primate Hippocampus CA1 region (Hippocampus CA1) [51], and GSE17757 included primate and human SFG [50].
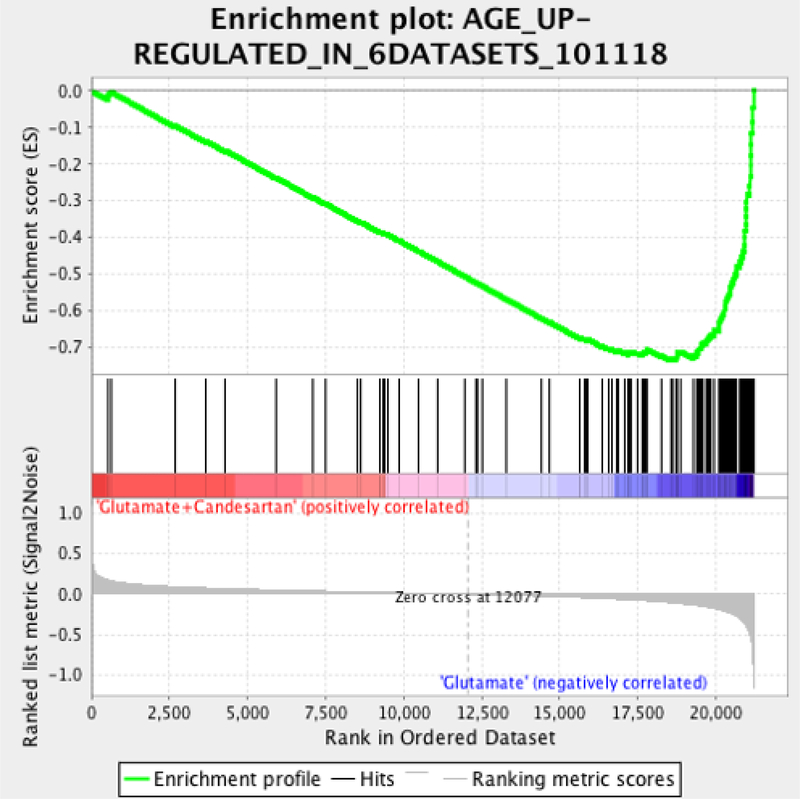
We defined 116 genes consistently up-regulated in all 6 datasets from aged brains by at least 1.2 folds (Online Resource 3_Age up-regulated in 6 datasets). Of these 116 aged brain genes, 99 genes were present in our neuronal array study. Of those 99 genes, 70 genes were up-regulated by glutamate (p-value<0.01) and down-regulated or normalized by Candesartan in our study. (Online Resource 3: GSEA AGE_UP-REGULATED_6DATASE). Moreover, when we used GSEA to run these 116 genes comnmonly up-regulated by aging over our entire neuronal microarray, we found a highly significant enrichment with genes up-regulated by glutamate cytotoxicity and down-regulated by Candesartan.(Fig 2 and Online Resource 3_Age up-regulated in 6 datasets). This is highly significant, considering that these results have been obtained from different tissues, different array platforms and different species.
Fig 2. Negative Correlation of Candesartan effect with common senescence genesets.
Gene Set Enrichment Analysis plots showing the negative correlations of genes up-regulated by glutamate and down-regulated by Candesartan in our neuronal culture with a geneset of common consistently up-regulated genes in 6 different brain aging datasets including different brain areas (hippocampus, entorhinal cortex, superior frontal gyrus) (See Online Resource 3). Data were taken from: GSE11882_Aging (Berchtold et al., 2008, https://www.ncbi.nlm.nih.gov/pubmed/18832152), GSE11697_HippaCA1_Aging (Blalock et al., 2010, https://www.ncbi.nlm.nih.gov/pubmed/20427664) and GSE17757_SFG_Aging (Somel et al., 2010, https://www.ncbi.nlm.nih.gov/pubmed/20647238).
Interestingly, two of the genes up regulated by glutamate in the 6 aging datasets and reversed by Candesartan encode for glutamate transporters and play a role in excitatory signaling. SLC25A18 is a mitochondrial glutamate transporter [67] and SLC7A11, encoding for the cystine/glutamate transporter xCT, has been found upregulated in Alzheimer’s disease patients [68].
To define the molecular pathways associated with these common aging geneset we used the DAVID Gene Onthologies. Using GO_BP, we identified 39 biological processes significantly associated with our findings including extracellular matrix disassembly, integrin-mediated signaling pathway, negative regulation of apoptotic process and inflammatory response (Online Resource 3, GO_BP). To identify key genes and pathways we used GOTERM_MF_DIRECT. We identified 17 associated pathways. Our results included receptor activity, cadherin binding involved in cell-adhesion and protein binding (Online Resource 3, GO_MF). Similar significant associations were found by GO_CC analysis of cellular components, including extracellular exosome, cell surface, plasma membrane and blood microparticles (Online Resource 3, GO_CC). Using Go_Diseases, we found 13 types of associated disorders. The most significantly associated diseases were immune, cardiovascular and pharmacogenomic disorders (Online Resource 3, GO_Diseases). To discover gene-disease associations, we compared our data base with gene ontology. We identified 19 GO_GAD diseases, including Type2 diabetes, asthma, systemic lupus erythematosus and chronic renal failure (Online Resource 3, GO_GAD_Disease).
We then used the Ingenuity Pathways Analysis (IPA) upstream regulator analysis for both the genes upregulated by glutamate and downregulated or normalized by Candesartan and the genes upregulated in 6 different aged brain. The list of the 116 genes most upregulated in 6 aging datasets and the list of the genes upregulated by Glutamate and downregulated by Candesartan (525 genes) were put in the IPA upstream regulators to determine if the same pathways were shared between aging and Candesartan action. We were able to show that both datasets share over 90% of their upstream regulators and that the z-scores for these common age-related upstream regulators are reversed when the CGC neurons injured by glutamate are treated with Candesartan (Online Resource 4: table IPA_ Combined GSE11882_CGC-Candesartan). This application yielded transcription regulators that are known from the literature to target the genes associated with aging and general inflammation and compared their z-score direction to weight in the predicted effect of the regulator on a subset of the genes (Table 2).
Table 2: Representative IPA upstream regulator’s z-score and p-value for aging up-regulated genes and CGC Glutamate-up/Candesartan-down regulated genes.
The table includes the z-scores and p-values for GSE11882_Aging_UP and for CGC_Glutamate-UP_Candesartan-Down.
| IPA UP- stream regulators |
z-score for GSE11882_ Aging_UP |
p-value for GSE11882_Aging_ UP |
z-score for CGC_ glutamate-UP_ Candesartan- Down |
p-value for CGC_glutamate- UP_Candesartan- Down |
|---|---|---|---|---|
| resveratrol | −1.618 | 1.49E-06 | 2.411 | 2.97E-20 |
| TNF | 7.042 | 3.26E-49 | −9.366 | 7.93E-71 |
| IL6 | 5.686 | 2.02E-22 | −6.103 | 3.12E-36 |
| Insulin | 1.879 | 4.78E-07 | −1.198 | 7.21E-08 |
| SIRT1 | −2.697 | 5.43E-06 | 2.004 | 1.89E-09 |
| FOXO1 | 3.896 | 1.08E-07 | −4.695 | 2.89E-17 |
| FOXO3 | 2.856 | 1.65E-08 | −4.011 | 5.92E-19 |
| TP53 | 4.253 | 5.97E-24 | −4.793 | 4.07E-26 |
| NFkB (complex) | 6.52 | 2.72E-14 | −7.381 | 8.12E-47 |
| IGF1 | 4.264 | 8.11E-10 | −5.135 | 4.00E-28 |
| TGFB1 | 7.112 | 7.65E-29 | −8.078 | 1.70E-60 |
| IL1B | 6.36 | 2.58E-31 | −8.866 | 1.32E-53 |
| PI3K (complex) | 3.979 | 1.49E-06 | −4.606 | 8.16E-18 |
| P38 MAPK | 3.876 | 2.19E-12 | −5.777 | 6.24E-31 |
| SMAD3 | 3.883 | 6.84E-08 | −4.574 | 1.59E-15 |
| SMAD4 | 2.318 | 4.54E-09 | −2.852 | 3.20E-18 |
| Mek | −0.156 | 8.47E-05 | −3.35 | 6.10E-23 |
| HRAS | 1.824 | 4.56E-24 | −2.673 | 8.01E-33 |
| ERK1/2 | 3.268 | 1.89E-08 | −4.736 | 3.74E-23 |
| PTEN | −1.579 | 6.83E-13 | 2.116 | 7.38E-16 |
| TSC2 | −4.303 | 4.49E-13 | 3.949 | 3.35E-16 |
| MAPK14 | 3.33 | 2.76E-06 | −4.292 | 1.40E-20 |
| JUN | 4.161 | 2.16E-11 | −3.877 | 2.14E-35 |
| FOS | 1.686 | 8.25E-09 | −3.339 | 2.46E-28 |
Many upstream-regulators associated with general inflammation pathways such as Il-6, TNF, the NFkB complex, insulin, TGFβ-SMAD and P38 MAPK-MEK-ERK1/2 show a positive z-score in aging, meaning that these upstream regulators can have a positive regulation (upregulation) on many downstream genes. These z-scores are then reversed for the same upstream regulators for the glutamate-up/Candesartan-down regulated genes. Reciprocally, resveratrol, a known anti-inflammatory and anti-aging compound and its positive partners SIRT1, TSC2 and PTEN [69–71], show a positive z-score for the glutamate-up/Candesartan-down regulated genes and negative z-score for aging. (Online Resource 4: Table IPA_ Combined GSE11882_CGC-Candesartan, Representative upstream regulators; Table 2).
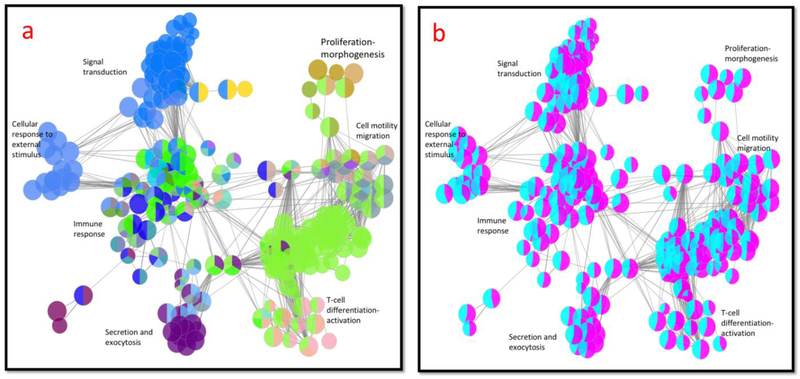
To further demonstrate the common pathways shared between these two genelists, we used the CluePedia plugin of Cytoscape (v3.7.1) [53]. As expected, these two lists show many pathways associated with signal transduction, cellular response to external stimuli, immune response, secretion and exocytosis, T-cell differentiation and activation, cell motility migration and proliferation-morphogenesis. (Fig 3 a). Then we asked the question of how many genes from each list are associated with each Gene Ontology pathway. Fig 3 b shows that each pathway shares 40–60% genes from each gene list. This mean that aging and glutamate cytotoxicity normalized by Candesartan share almost the same pathways.
Fig 3. Pathways associated with differentially expressed genes.
Pathways associated with differentially expressed genes in 4 aging brain regions from GSE11882 (1.2 Fold change and p-value<0.05) and genes differentially expressed between rat cortical granule cells treated with cytotoxic glutamate versus cells pre-treated with Candesartan and then glutamate (1.2 Fold change and p-value<0.05). Figures were generated side-by-side using the CluePedia plugin of Cytoscape (v3.7.1) (http://www.cytoscape.org/). a: Color coded network of gene ontology pathways and their connection. b: The same network as in a. but this time showing the pie distribution of genes up-regulated in aging (purple) and genes up-regulated by glutamate and down-regulated by Candesartan(magenta).
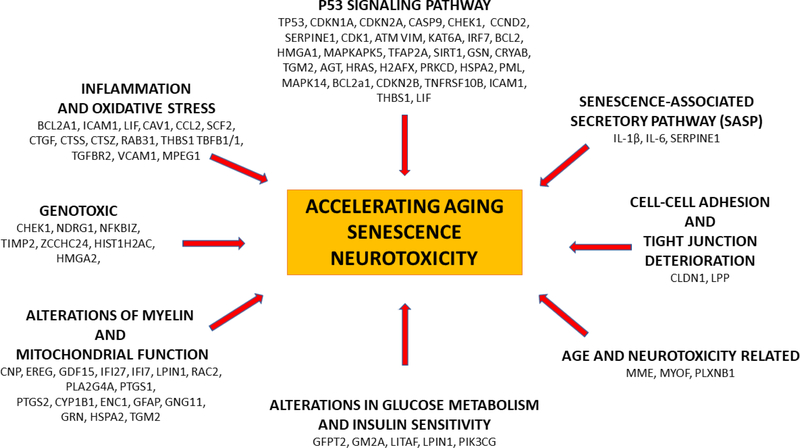
Using Gene Set Enrichment Analysis (GSEA), over 70 genes significantly upregulated by glutamate in our neuronal culture dataset and normalized by exposure to Candesartan are enriched in several aging and senescence data sets. These genes are involved in all the major pathways that are considered hallmarks of aging and senescence, (Online Resource 5) (Fig 4). The defined hallmarks of aging are closely interrelated, and many genes play essential roles in several major signaling systems. For example, telomere dysfunction alters mitochondrial function, enhances oxidative stress and upregulates the inflammasome. abnormality, oxidative stress, and hyperactivation of the NLRP3 inflammasome [72]. Many genes, including those encoding for transforming growth factor beta (TGFβ) or tumor necrosis factor-alpha (TNFα) exert pro-longevity and anti-longevity effects, and their activity may enhance neuroprotection or promote neurodegenerative disorders, depending on the biological and environmental context [73].
Fig 4. Influence of Candesartan on genes associated with hallmarks of senescence.
The Fig represents principal hallmarks of senescence and associated genes upregulated by glutamate in our database, normalized by Candesartan and enriched in selected datasets (Figs 1 and 2, Online Resource 3). Many genes associate and influence each other.
A pathway of major importance in aging is that controlled by TP53. The TP53 gene encodes the tumor protein p53, regulating the cell cycle arrest, mediates DNA damage leading to cell senescence, cell death and many other functions. Unrestrained and excessive p53 activation is detrimental to healthy aging [74–77]. p53 is activated by multiple stressors, including glutamate excitotoxicity [78]. Nine genes from these studies are part of the p53 signaling pathway (TP53, CDKN1A, CDKN2A, CASP9, CHEK1, CCND2, SERPINE1, CDK1 and ATM (Table 3, Fig 4) and 24 genes have a protein interaction with p53 protein (CDKN2A, VIM, KAT6A, IRF7, BCL2, CHEK1, HMGA1, MAPKAPK5, TFAP2A, SIRT1, GSN, CRYAB, TGM2, MAPK14, AGT, HRAS, H2AFX, PRKCD, CDK1, ATM, HSPA2, TP53 and PML) (Table 3, Online Resource 5; Fig 4)
Table 3. p53-associated genes and dataset enrichment correlation.
The table includes genes associated to the p53 pathway that are enriched in analyzed datasets, including gene symbols, corresponding encoding protein and dataset enrichment correlation
| Gene symbol | Encoded protein | Dataset enrichment correlation |
|---|---|---|
| TP53 | tumor protein p53 | FRIDMAN_SENESCENCE_UP GO_AGING GO_CELL_AGING GO_CELLULAR_SENESCENCE |
| CDKN1A | cyclin dependent kinase inhibitor 1A | FRIDMAN_SENESCENCE_UP GO_AGING GO_CELL_AGING GO_CELLULAR_SENESCENCE GSE28464_IMR90CELLS_RAS_SENESCENCE_UP KAYO_AGING_MUSCLE_UP TANG_SENESCENCE_TP53_TARGETS_UP |
| CDKN2A | cyclin dependent kinase inhibitor 2A | FRIDMAN_SENESCENCE_UP GO_AGING GO_CELL_AGING GO_CELLULAR_SENESCENCE GSE28464_IMR90CELLS_RAS_SENESCENCE_UP |
| CASP9 | caspase 9 | GO_AGING GSE40349_IMR90_RAS_SENECENCE_UP Reiter E_GSE8150_NEOCORTEX_AGING UP |
| CHEK1 | checkpoint kinase 1 | GO_AGING GO_CELL_AGING KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_UP |
| CCND2 | cyclin D2 | GSE40349_IMR90_RAS_SENECENCE_UP RODWELL_AGING_KIDNEY_UP TANG_SENESCENCE_TP53_TARGETS_UP |
| SERPINE1 | serpin family E member 1 | FRIDMAN_SENESCENCE_UP GO_AGING GO_CELL_AGING KAYO_AGING_MUSCLE_UP |
| CDK1 | cyclin dependent kinase 1 | GO_AGING GO_CELL_AGING KYNG_WERNER_SYNDROM_AND_NORMAL_AGING_UP |
| ATM | ATM serine/threonine kinase | GO_AGING GO_CELL_AGING GSE28464_IMR90CELLS_RAS_SENESCENCE_UP |
Members of the p53 pathway include genes reported to be enhanced in by inflammatory stimuli, in aging and in Alzheimer’s disease: BCL2A1, encoding BCL-2-related protein A, a direct transcription target of NF-κB in response to inflammatory mediators [79], CDKN1A and CDKN2B, encoding for p21, a cyclin-dependent kinase inhibitor protein, a major target of p53 associated to DNA damage and a senescence associated gene [80–83] (Table 3). CHEK1, encoding for the CHK1 checkpoint homolog, increased in aging and Alzheimer’s disease [84], TNFRSF10B encoding for the tumor necrosis factor receptor superfamily member 10B or death receptor 5 (DR5) that mediates age-related p53-induced apoptosis [85]; ICAM1, encoding for the intracellular adhesion molecule 1, participating in the inflammatory phenotype of senescent cells [86], MAPK14, encoding for the mitogen-activated kinase 14, is activated by environmental stressors and pro-inflammatory cytokines, regulates p53 inducing senescence, plays a key role in stress factors affecting genomic integrity, and participates in Alzheimer’s disease [87, 88], THBS1, encoding for thrombospondin 1, leads to enhanced ROS and increased p53 transcription with advancing age [89–91], LIF, encoding for the leukemia inhibitory factor, a protein regulated by p53, stimulated by pro-inflammatory cytokines and with a major role in inflammation [92], and TFAP2A, encoding for the transcription factor AP-2 alpha and associated with DNA hypermethylation that directly interacts with p53 [93, 94] (Fig 4).
Within this pathway, SERPINE1, encoding for the plasminogen activator inhibitor-1 (PAI-1) [95, 96] is of great interest. PAI-1 is the major inhibitor of endogenous thrombolysis, promoting thrombosis, it is strongly activated by Angiotensin II [97], and inhibited by ARBs, including Candesartan [98]. SERPINE1 is both a marker and a mediator of senescence and many age-related disorders [99–103] and it is much higher in p16-positive senescence cells [104]. It is activated by ROS, oxidative stress, the DNA Damage Response (DDR), the pro-senescence TGFβ, glutamate excitotoxicity and stress-induced anxiety-like behavior [105, 106].
Inflammation and oxidative stress are major determinant of aging and senescence. Genes involved in inflammatory pathways include BCL2A1, ICAM-1, LIF, CAV1 encoding calveolin 1, upregulated by oxidative stress [107], CCL2, encoding chemokine (C-C motif) ligand 2, associated with neurodegenerative disorders [108]; CSF2, encoding the colony stimulating factor 2, part of the inflammatory cascade controlled by NF-κB [109], CTGF, encoding for connective tissue growth factor, induced by oxidative stress and in accelerating aging models [110], CTSS, encoding cathepsin S, associated with olfactory dysfunction [111] and CTSZ, encoding for Cathepsin Z involved in dopamine neuron death [112], and RAB31, encoding for Ras-related protein Rab-31, stimulating high levels of oxidative stress [113] and THBS1, encoding for thrombospondin 1, a protein enhancing ROS and contributing to coronary ischemia in advancing age [90].
TGFβ also contributes to cellular senescence. TGFB1I1, encoding for transforming growth factor beta-1-induced transcript 1 protein, is involved in multiple functions including apoptosis, the immune system and increasing IL-1-beta, and TGFBR2, encoding for transforming growth factor, beta receptor II is a TGF beta receptor increased in rodent models and in patients with Alzheimer’s Disease linking neuroinflammation and amyloidosis [114] (Online Resource 5).
Some additional inflammatory genes are upregulated by Angiotensin II and reduced by AT1 receptor blockade, such as VCAM-1 encodes the cell adhesion molecule vascular cell adhesion protein 1. Its production is enhanced in response to tumor necrosis factor-alpha (TNF-α) and interleukin-1 (IL-1), that is involved in high-glucose endothelial cell senescence, [115, 116], and MPEG1, encoding for the macrophage expressed gene 1 [117] an inflammatory gene that reduces necessary angiogenic responses to ischemia in aged rats [118].
Candesartan normalizes the glutamate-upregulated Senescence-Associated Secretory Pathway (SASP). The SASP increases in multiple tissues with aging and is triggered by senescence cells experiencing DNA damage [119], and Candesartan normalizes the upregulated SASP as a consequence of glutamate toxicity. Normalized genes include including IL1β, encoding for the pro-inflammatory cytokine interleukin 1β, [120, 121] and IL6, encoding for IL-6, that stimulates inflammation and auto-immune processes in multiple age-related diseases [122, 123] and SERPINE1.
Abnormal expression of a number of genes included in our analysis is genotoxic. They are directly involved in DNA alterations leading to genome instability during aging, and in particular telomere dysfunction, leading to mitochondrial abnormality, oxidative stress and hyperactivation of the NLRP3 inflammasome. These genes include CHEK1 [84], NDRG1, encoding for the NDRG1 protein, strongly associated with age and disorders of DNA methylation and with multiple functions in stress responses, immunity myelination and cell adhesion [124, 125], NFKBIZ encoding for NF-kappa-B inhibitor zeta a nuclear protein induced by inflammation and TNF-α, associated with senescence and cell death [126–129], and TIMP2, encoding for the tissue inhibitor of metalloproteinases 2 (TIMP2), increasing after genotoxic stress and part of the SASP [130]. ZCCHC24 encodes for zinc finger, CCHC domain containing 24. This protein binds nucleic acids and is upregulated in aging, contributing to genomic instability and telomere attrition [7, 131, 132]. Other genotoxic-related genes include those encoding histones such as HIST1H2AC, encoding for histone 1, H2ac, a protein fundamental for DNA function [7, 104, 133], HMGA2, encoding for high mobility group AT-hook 2, promoting stem cell aging [134] (Online Resource 5).
Genes with roles in cell-cell adhesion and tight junctions include CLDN1 encodes for Claudin-1, involved in tight junction deterioration [135] and LPP, encoding for LIM domain containing preferred translocation partner in lipoma or lipoma-preferred partner, involved in cell-cell adhesion and cell motility [136] (Online Resource 5).
Genes involved in myelin and mitochondrial function and enhanced during aging include CNP, encoding for 2’,3’-cyclic nucleotide 3’phosphodiesterase [137, 138], EREG, encoding for epiregulin [139], GDF15, encoding for growth differentiation factor 15, a protein that contributes to radiation-induced senescence mediated by the p16 pathway [140, 141], IFI27, encoding for interferon, alpha-inducible protein 27, a protein that destabilizes mitochondrial membrane function sensitizing cells to apoptotic stimuli [142, 143], IRF7, encoding for interferon regulatory factor 7, involved in the transcriptional activation of virus-induced cellular genes, and increased expression of interferon type 1, an mRNA signature in the aging brain [144–148], LPIN1 and RAC2 encoding for Ras-related C3 botulinum toxin substrate 2, associated with decline in mitochondrial respiration and prostaglandin metabolism, that delays post-stroke angiogenesis in the aging brain [117].
Alterations in prostaglandin metabolism play significant roles in aging and the development of neurodegenerative disorders, including alterations in expression of PLA2G4A, encoding for Cytosolic phospholipase A2, associated with decline in mitochondrial respiration in the female aging brain and Alzheimer’s disease [149], PTGS1, encoding for Cyclooxygenase 1 (COX-1), prostaglandin-endoperoxide synthase 1, increased in older subjects with schizophrenia [150] and PTGS2 encoding for prostaglandin-endoperoxide synthase 2, cyclooxygenase-2 or COX-2, involved in premature aging [151].
Additional senescence-associated genes include CYP1B1, encoding for Cytochrome P450, family 1, subfamily B, polypeptide 1 [152], ENC1, encoding for ectodermal-neural cortex (with BTB-like domain) with a proposed role in the cognitive decline during aging [153], GFAP, encoding for the glial fibrillary acidic protein, enhanced in models of Alzheimer’s disease and a marker of neurological damage [154], GNG11, encoding for guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-11 and inducing cellular senescence [155], . GRN, encoding for granulin and associated with hippocampal sclerosis [156, 157], HSPA2, encoding for heat shock 70kDa protein 2 that has been associated with increased amyloid-beta and tau in Alzheimer’s disease [158]. TGM2 encodes for tissue transglutaminase, a protein with a role in apoptosis and inflammation that increases resistance to proteolysis resulting in abnormal aggregation of proteins and involved in Alzheimer’s and Parkinson’s disease [159–162] (Online Resource 5). MME encodes for membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10) or neprilysin. This protein is associated with Alzheimer’s disease and renal damage; it is a downstream effector of PI3K mediating the induction of senescence, and a compound combining neprilysin and AT1 receptor blockade is beneficial for the treatment of heart failure [163, 164]. MYOF, encoding for fer-1-like 3, myoferlin, that has been associated with age in human populations [165] and PLXNB1, encoding for Plexin B1 and associated with cognitive decline and amyloid neuropathology [166] (Online Resource 5).
A number of genes that regulate glucose metabolism and insulin sensitivity, with enhanced expression in aging and diabetes, include GFPT2, encoding for glutamine-fructose-6-phosphate transaminase and associated with type2 diabetes [167], GM2A, encoding for GM2 ganglioside activator and enhancing insulin resistance [168], LITAF, encoding for the lipopolysaccharide-induced TNF factor, enhanced in diabetes, aging and inflammatory processes [169], LPIN1, encoding for lipin 1, a protein associated with insulin resistance, inflammation and myelin disease [170] and PIK3CG, encoding for phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma, increasing oxidative stress and upregulating MAPK, playing a role in Angiotensin II-induced NADPH oxidase activation and leading to cardiomyopathy through AT1 receptor activation, that is downregulated by AT1 receptor inhibition [171, 172] (Online Resource 5).
The expression of some genes, upregulated by glutamate and normalized by Candesartan, may represent compensatory mechanisms to injury. For example, overexpression of MCTP1, encoding for multiple C2 domains, transmembrane 1, inhibits oxidative stress produced by glutamate excitotoxicity [173]. NRG1 encodes for Neuroregulin 1, a protein that attenuates stress-induced vascular senescence [174]. TGIF1, encoding for Homeobox protein TGIF1, binds to the retinoid X receptor responsive element. When expression is reduced, TGFβ signaling is increased leading to DNA damage and premature senescence [175, 176], and TNFAIP3, encoding for tumor necrosis factor, alpha-induced protein 3, a protein induced by tumor necrosis factor that inhibits TNF-mediated apoptosis; it is critical for limiting inflammation by terminating TNF-induced NF-κ B responses and critical for the homeostatic role of telomeres [72] (Online Resource 5).
4. CONCLUSIONS
Our initial global gene analysis of our neuronal culture revealed that multiple alterations in gene expression resulting from glutamate excitotoxicity were reversed or normalized by incubation in the presence of the ARB Candesartan, and that these alterations in gene expression significantly correlated with findings in autopsy brains from patients with Alzheimer’s disease. We had now compared our findings with those reported in multiple aging and senescence datasets, and we found significant correlations with alterations in expression of multiple genes reported in aging and senescence datasets.
In our study we used a culture of primary neurons, the cerebellar granule cells, established as the best model to determine mechanisms of neuronal survival, apoptosis and aging [177–180]. In addition, our results support the hypothesis of the importance of the cerebellum in cognitive behavior and its influence in the aging process [181–183].
We found enriched alterations in gene expression related to all major mechanisms associated with aging and senescence, including defects of the DNA-repair and telomere maintenance systems, the tumor suppressor pathway, reduction of synaptic plasticity, vesicular transport and mitochondrial function, alterations in glucose metabolism, excessive oxidative stress and inflammation, and the SASP. These alterations have been associated with reduction on neuronal survival, decreased learning and memory characteristic of neurodegenerative and many other age-related disorders, and common not only for brain disorders but for age-related diseases of the whole organism. Interestingly, most of the molecules that influence the phenotypic changes of aging also regulate cellular senescence, suggesting a causative link between cellular senescence and aging.
Our results support the hypothesis of a major negative, pro-aging and pro-senescence, influence of excessive Angiotensin II AT1 receptor activity not only in the brain but also in the periphery, and demonstrate the overall protective, anti-aging effects of ARB administration.
Supplementary Material
Whole Gene Set Enrichment Analysis output for all aging associated genesets with plots, statistics, gene status and heatmaps. GO: Gene Ontology Data Archive. Genesets with links and References are listed in Table 1. :
The table shows the whole Gene Set Enrichment Analysis output for all senescence associated genesets with plots, statistics, gene status and heatmaps. GO: Gene Ontology Data Archive. Genesets with links and References are listed in Table 1.
We looked for age upregulated genes in 6 different brain aging datasets (GSE11882_PCG, GSE11882_SFG, GSE11882_Hippo, GSE11882_EnthorinalCrtx, GSE11697_HippoCA1 and GSE17757_SFG). 116 genes were consistently up-regulated by 1.2 folds in all 6 different brain tissues aging datasets (blue columns) and 99 of them are also present in the CGC dataset (Green columns). Of the 99 genes upregulated in aging and present in the CGC study, 70 of them are also upregulated by Glutamate alone and are down-regulated or normalized by Candesartan treatment. In each comparison dataset, the p-value, Fold Change and change direction are represented. Aging Data are taken from GSE11882, GSE11697 and GSE17757 (see detailed information in Table 1). GoRilla analysis is presented in this order: GoRilla_process GoRilla_Function, GoRilla_Component (description, p-value, FDR q-value, enrichment, and genes), GO_BP, GO_MF, GO_CC, GO_diseases, GO_GAD_Disease (category, term, RT, genes, count, %, p-value, Benjamini).
The list of all the IPA up-stream regulators for the aging associated geneset (grey rows, 1101 regulators) and the CGC glutamate up-regulated and Candesartan down-regulated geneset (yellow rows, 1203 regulators) were sorted based on the upstream regulator. The regulator that are unique to the CGC glutamate+Candesartan are labeled in red and the ones unique to the aging dataset are in blue. The table includes their predicted activation state, the activation z-score, the p-value of overlap, the target molecules and whether the upstream regulators are common or unique, referring to regulators that are common in both genesets or unique to aging (blue label) or unique to Glutamate/Candesartan (red label). The negative z-score is associated with predicted inhibition of the upstream regulator and the positive z-score is associated with predictive activation of the upstream regulator (Combined upstream regulators). A list of 28 representative upstream regulators based on known and published genes, compounds and pathways associated with aging includes z-scores and p-values for GSE11882_Aging_UP and for CGC_Glutamate-UP_Candesartan-Down that are upregulated consistently in 6 different aging datasets (Table 2).
List of genes that are GSEA enriched in at least 3 out of 19 aging and senescence datasets from different tissues and cells studied.
Acknowledgements.
The authors wish to thank the Comparative genomics and Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA (AGE) and the Department of Pharmacology and Physiology, Georgetown University Medical Center, Washington, DC 20057, USA (JMS) for their support during the preparation of this manuscript. AGE was supported by the Intramural NHGRI. JMS did not receive any support for this study. The funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Abbreviations
- ANOVA
ANalysis Of Variance
- ARBs
Angiotensin Receptor Blockers
- CGC
cerebellar granule cells
- COX-1
cyclooxygenase 1
- COX-1
cyclooxygenase-2
- GEO
Gene Expression Omnibus
- GO
Gene Ontology database
- GSEA
Gene Set Enrichment Analysis
- IL-1
interleukin-1
- IPA
Ingenuity pathway analisis
- MSigDB
Broad Molecular Signatures Database v5.0
- NCBI
National Center for Biotechnology Information
- PAI-1
plasminogen activator inhibitor
- PCG
posterior cingulate cortex
- RMA
Robust Multichip Average
- SASP
Senescence-Associated Secretory Pathway
- SFG
superior frontal gyrus
- TGFβ
transforming growth factor beta
- TNFα
tumor necrosis factor-alpha
Footnotes
Compliance with Ethical Standards
Conflict of Interest: The authors declare that they have no conflict of interest.
REFERENCES
- [1].Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O’Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T, Inflammation and Alzheimer’s disease. Neurobiol Aging. (2000) 21:383–421. [PubMed: 10858586] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Grammas P, Martinez J, Miller B, Cerebral microvascular endothelium and the pathogenesis of neurodegenerative diseases. Expert Rev Mol Med. (2011) 13:e19. 10.1017/S1462399411001918 [DOI] [PubMed] [Google Scholar]
- [3].Hanke ML, Kielian T. Toll-like receptors in health and disease in the brain: mechanisms and therapeutic potential. Clin Sci. 2011; 121:367–387. https://doi:10.1042/CS20110164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lartaud I, Makki T, Bray-des-Boscs L, Niederhoffer N, Atkinson J, Corman B, Capdeville-Atkinson C, Effect of chronic Ang I-converting enzyme inhibition on aging processes. IV Cerebral blood flow regulation. Am J Physiol. (1994) 267:R687–R694. 10.1152/ajpregu.1994.267.3.R687 [DOI] [PubMed] [Google Scholar]
- [5].Letiembre M, Hao W, Liu Y, Walter S, Mihaljevic I, Rivest S, Hartmann T, Fassbender K, Innate immune receptor expression in normal brain aging. Neuroscience. (2007)146:248–254. 10.1016/j.neuroscience.2007.01.004 [DOI] [PubMed] [Google Scholar]
- [6].Letiembre M, Liu Y, Walter S, Hao W, Pfander T, Wrede A, Schulz-Schaeffer W, Fassbender K, Screening of innate immune receptors in neurodegenerative diseases: a similar pattern. Neurobiol Aging. (2009) 30:759–768. 10.1016/j.neurobiolaging.2007.08.018 [DOI] [PubMed] [Google Scholar]
- [7].López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging. Cell. (2013)153:1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miller KR, Streit WJ, The effects of aging, injury and disease on microglial function: a case for cellular senescence. Neuron Glia Biol. (2007) 3:245–253. 10.1017/S1740925X08000136 [DOI] [PubMed] [Google Scholar]
- [9].Wolkowitz OM, Epel ES, Reus VI, Mellon SH, Depression gets old fast: do stress and depression accelerate cell aging? Depression Anxiety. (2010) 27:327–338. 10.1002/da.20686 [DOI] [PubMed] [Google Scholar]
- [10].Wyss-Coray T, Ageing, neurodegeneration and brain rejuvenation. Nature. (2016) 539:180–186. 10.1038/nature20411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saavedra JM, Angiotensin II AT(1) receptor blockers as treatments for inflammatory brain disorders. Clin Sci (Lond). (2012a)123:567–590. 10.1042/CS20120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Saavedra JM, Angiotensin II AT(1) receptor blockers ameliorate inflammatory stress: a beneficial effect for the treatment of brain disorders. Cell Mol Neurobiol. (2012b) 32:667–681. 10.1007/s10571-011-9754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Saavedra JM, Beneficial effects of Angiotensin II receptor blockers in brain disorders. Pharmacol Res. (2017)125:91–103. 10.1016/j.phrs.2017.06.017 [DOI] [PubMed] [Google Scholar]
- [14].Saavedra JM, Benicky J, Brain and peripheral angiotensin II play a major role in stress. Stress. (2007)10:185–193. 10.1080/10253890701350735 [DOI] [PubMed] [Google Scholar]
- [15].Basso N, Paglia N, Stella I, de Cavanagh EM, Ferder L, del Rosario Lores Arnaiz M, Inserra F, Protective effect of the inhibition of the renin-angiotensin system on aging. Regul Pept. (2005) 128:247–252. 10.1016/j.regpep.2004.12.027 [DOI] [PubMed] [Google Scholar]
- [16].de Cavanagh EM, Inserra F, Ferder L, Angiotensin II blockade: how its molecular targets may signal to mitochondria and slow aging. Coincidences with calorie restriction and mTOR inhibition. Am J Physiol Heart Circ Physiol (2015)309:H15–H44. 10.1152/ajpheart.00459.2014 [DOI] [PubMed] [Google Scholar]
- [17].Villapol S, Saavedra JM, Neuroprotective effects of angiotensin receptor blockers. Am J Hypertens. (2015)28:289–299. 10.1093/ajh/hpu197 [DOI] [PubMed] [Google Scholar]
- [18].Benicky J, Sánchez-Lemus E, Honda M, Pang T, Orecna M, Wang J, Leng Y, Chuang DM, Saavedra JM, Angiotensin II AT1 receptor blockade ameliorates brain inflammation. Neuropsychopharmacology. (2011) 36:857–870. 10.1038/npp.2010.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Elkahloun AG, Hafko R, Saavedra JM, An integrative genome-wide transcriptome reveals that candesartan is neuroprotective and a candidate therapeutic for Alzheimer’s disease. Alzheimers Res Ther. (2016) 8:5 10.1186/s13195-015-0167-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Elkahloun AG, Rodriguez Y, Alaiyed S, Wenzel E, Saavedra JM, Telmisartan Protects a Microglia Cell Line from LPS Injury Beyond AT1 Receptor Blockade or PPARγ Activation. Mol Neurobiol. (2019) 56:3193–3210. 10.1007/s12035-018-1300-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang J, Pang T, Hafko R, Benicky J, Sanchez-Lemus E, Saavedra JM, Telmisartan ameliorates glutamate-induced neurotoxicity: roles of AT(1) receptor blockade and PPARγ activation. Neuropharmacology. (2014)79:249–261. 10.1016/j.neuropharm.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gilliam-Davis S, Gallagher PE, Payne VS, Kasper SO, Tommasi EN, Westwood BM, Robbins ME, Chappell MC, Diz DI, Long-term systemic angiotensin II type 1 receptor blockade regulates mRNA expression of dorsomedial medulla renin-angiotensin system components. Physiol Genomics. (2011) 43):829–835. 10.1152/physiolgenomics.00167.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trofimiuk E, Wielgat P, Braszko JJ, Candesartan, angiotensin II type 1 receptor blocker is able to relieve age-related cognitive impairment. Pharmacol Rep. (2018) 70:87–92. 10.1016/j.pharep.2017.07.016 [DOI] [PubMed] [Google Scholar]
- [24].Baiardi G, Bregonzio C, Jezova M, Armando I, Saavedra JM, Angiotensin II AT1 Receptor Blockade Prolongs the Lifespan of Spontaneously Hypertensive Rats and Reduces Stress-Induced Release of Catecholamines, Glucocorticoids, and Vasopressin. Ann N Y Acad Sci. (2004) 1018:131–136. 10.1196/annals.1296.015 [DOI] [PubMed] [Google Scholar]
- [25].Basso N, Cini R, Pietrelli A, Ferder L, Terragno NA, Inserra F. Protective effect of long-term angiotensin II inhibition. Am J Physiol Heart Circ Physiol. (2007) 293:H1351–H1358. 10.1152/ajpheart.00393.2007 [DOI] [PubMed] [Google Scholar]
- [26].Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G, Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. (2009)119:524–530. 10.1172/JCI36703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bennai F, Morsing P, Paliege A, Ketteler M, Mayer B, Tapp R, Bachmann S, Normalizing the expression of nitric oxide synthase by low-dose AT1 receptor antagonism parallels improved vascular morphology in hypertensive rats. J Am Soc Nephrol. (1999)10 Suppl 11:S104–S15. PMID: 9892150 [PubMed] [Google Scholar]
- [28].Linz W, Heitsch H, Schölkens BA, Wiemer G, Long-term angiotensin II type 1 receptor blockade with fonsartan doubles lifespan of hypertensive rats. Hypertension. (2000) 35:908–913. http://www.ncbi.nlm.nih.gov/pubmed/10775560 [DOI] [PubMed] [Google Scholar]
- [29].Barger JL, Vann JM, Cray NL, Pugh TD, Mastaloudis A, Hester SN, Wood SM, Newton MA, Weindruch R, Prolla TA, Identification of tissue-specific transcriptional markers of caloric restriction in the mouse and their use to evaluate caloric restriction mimetics. Aging Cell. (2017) 16:750–760. 10.1111/acel.12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ando H, Zhou J, Macova M, Imboden H, Saavedra JM, Angiotensin II AT1 receptor blockade reverses pathological hypertrophy and inflammation in brain microvessels of spontaneously hypertensive rats. Stroke. (2004) 35:1726–1731. 10.1161/01.STR.0000129788.26346.18 [DOI] [PubMed] [Google Scholar]
- [31].Benicky J, Sánchez-Lemus E, Pavel J, Saavedra JM, Anti-inflammatory effects of angiotensin receptor blockers in the brain and the periphery. Cell Mol Neurobiol. (2009) 29:781–792. 10.1007/s10571-009-9368-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Danielyan L, Klein R, Hanson L, Buadze M, Schwab M, Gleiter CH, Frey WH, Protective effects of intranasal losartan in the APP/PS1 transgenic mouse model of Alzheimer disease. Rejuvenation Res. (2010)13:195–201. 10.1089/rej.2009.0944 [DOI] [PubMed] [Google Scholar]
- [33].Nakagawa T, Hasegawa Y, Uekawa K, Kim-Mitsuyama S, Chronic kidney disease accelerates cognitive impairment in a mouse model of Alzheimer’s disease, through angiotensin II. Exp Gerontol. (2017)87:108–112. 10.1016/j.exger.2016.11.012. [DOI] [PubMed] [Google Scholar]
- [34].Rodriguez-Perez AI, Dominguez-Meijide A, Lanciego JL, Guerra MJ, Labandeira-Garcia JL, Dopaminergic degeneration is enhanced by chronic brain hypoperfusion and inhibited by angiotensin receptor blockage. Age (Dordr). (2013);35:1675–1690. 10.1007/s11357-012-9470-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Saavedra JM, Evidence to Consider Angiotensin II Receptor Blockers for the Treatment of Early Alzheimer’s Disease. Cell Mol Neurobiol. (2016) 36:259–279. 10.1007/s10571-015-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou J, Ando H, Macova M, Dou J, Saavedra JM, Angiotensin II AT1 receptor blockade abolishes brain microvascular inflammation and heat shock protein responses in hypertensive rats. J Cereb Blood Flow Metab. (2005) 25:878–886. 10.1038/sj.jcbfm.9600082 [DOI] [PubMed] [Google Scholar]
- [37].Fogari R, Zoppi A, Effect of antihypertensive agents on quality of life in the elderly. Drugs Aging. (2004) 21:377–393. 10.2165/00002512-200421060-00003 [DOI] [PubMed] [Google Scholar]
- [38].Fournier A, Oprisiu-Fournier R, Serot JM, Godefroy O, Achard JM, Faure S, Mazouz H, Temmar M, Albu A, Bordet R, Hanon O, Gueyffier F, Wang J, Black S, Sato N, Prevention of dementia by antihypertensive drugs: how AT1-receptor-blockers and dihydropyridines better prevent dementia in hypertensive patients than thiazides and ACE-inhibitors. Exp Rev Neurother. (2009) 9:1413–1431. 10.1586/ern.09.89 [DOI] [PubMed] [Google Scholar]
- [39].Li NC, Lee A, Whitmer RA, Kivipelto M, Lawler E, Kazis LE Wolozin B, Use of angiotensin receptor blockers and risk of dementia in a predominantly male population: prospective cohort analysis. BMJ (2010) 340, b5465 10.1136/bmj.b5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zanchetti A, Elmfeldt D, Findings and implications of the Study on Cognition and Prognosis in the Elderly (SCOPE): a review. Blood Pressure. (2006) 15:71–79. 10.1080/08037050600771583 [DOI] [PubMed] [Google Scholar]
- [41].Gene Set enrichment analysis (GSEA): http://www.broadinstitute.org/gsea/
- [42].Broad Molecular Signatures Database v5.0 (MSigDB) http://www.broadinstitute.org/gsea/
- [43].Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC, PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics (2003)34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- [44].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. (2005) 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gene Omnibus database: http://www.ncbi.nlm.nih.gov/geo/
- [46].Reiter E, Jiang Q, Christen S, Anti-inflammatory properties of alpha- and gamma-tocopherol. Mol Aspects Med. (2007) 28:668–691. 10.1016/j.mam.2007.01.003.GSE8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Takebayashi S, Tanaka H, Hino S, Nakatsu Y, Igata T, Sakamoto A, Narita M, Nakao M, Retinoblastoma protein promotes oxidative phosphorylation through upregulation of glycolytic genes in oncogene-induced senescent cells. Aging Cell. (2015) 14:689–697. 10.1111/acel.12351GSE60652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavaré S, Inoki K, Shimizu S, Narita M, Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. (2011) 332:966–970. https://doi.org/10.1126/science.1205407GSE28464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW, Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. (2008) 105:15605–15610. https://doi.org/10.1073/pnas.0806883105GSE11882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, Hu H, Lachmann M, Zeng R, Chen W, Khaitovich P, MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. (2010) 20:1207–1218. 10.1101/gr.106849.110GSE17757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Blalock EM, Grondin R, Chen KC, Thibault O, Thibault V, Pandya JD, Dowling A, Zhang Z, Sullivan P, Porter NM, Landfield PW, Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J Neurosci. (2010) 30:6058–6071. 10.1523/JNEUROSCI.3956-09.2010GSE11697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].IPA Ingenuity pathway analysis. (http://www.ingenuity.com)
- [53]. CluePedia plugin of Cytoscape (v3.7.1) http://apps.cytoscape.org/apps/cluepedia.
- [54].Lee CK, Weindruch R, Prolla TA, Gene-expression profile of the ageing brain in mice. Nat Genet. (2000) 25:294–297. 10.1038/77046 [DOI] [PubMed] [Google Scholar]
- [55].Kyng KJ, May A, Kølvraa S, Bohr VA, Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc Natl Acad Sci U S A. (2003) 100:12259–12264. https://doi.org/10.1073/pnas.2130723100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].de Magalhães JP, Curado J, Church GM, Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. (2009) 25:875–881. 10.1093/bioinformatics/btp073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lee CK, Klopp RG, Weindruch R, Prolla TA, Gene expression profile of aging and its retardation by caloric restriction. Science. (1999) 285:1390–1393. 10.1126/science.285.5432.1390 [DOI] [PubMed] [Google Scholar]
- [58].Kayo T, Allison DB, Weindruch R, Prolla TA, Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc Natl Acad Sci U S A. (2001) 98:5093–5098. 10.1073/pnas.081061898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, Myers BD, Brooks JD, Davis RW, Higgins J, Owen AB, Kim SK, A transcriptional profile of aging in the human kidney. PLoS Biol. (2004) 2:e427. 10.1371/journal.pbio.0020427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA, Gene regulation and DNA damage in the ageing human brain. Nature. (2004)429:883–891. https://doi.org/10.1038/nature02661 [DOI] [PubMed] [Google Scholar]
- [61].Tran D, Bergholz J, Zhang H, He H, Wang Y, Zhang Y, Li Q, Kirkland JL, Xiao ZX, Insulin-like growth factor-1 regulates the SIRT1-p53 pathway in cellular senescence. Aging Cell. (2014) 13:669–678. 10.1111/acel.12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fridman AL, Tainsky MA, Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. (2008) 27:5975–5987. 10.1038/onc.2008.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tang X, Milyavsky M, Goldfinger N, Rotter V, Amyloid-beta precursor-like protein APLP1 is a novel p53 transcriptional target gene that augments neuroblastoma cell death upon genotoxic stress. Oncogene. (2007)26:7302–7312. 10.1038/sj.onc.1210542 [DOI] [PubMed] [Google Scholar]
- [64].Bunz F A, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B, Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. (1998) 282 (5393): 1497–1501. https://doi.org/10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- [65].Xu X, Kim JJ, Li Y, Xie J, Shao C, Wei JJ, Oxidative stress-induced miRNAs modulate AKT signaling and promote cellular senescence in uterine leiomyoma. J Mol Med (Berl). (2018) 96:1095–1106. 10.1007/s00109-018-1682-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD, Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. (2013)13:331–341. 10.1016/j.spinee.2012.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE, Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem. (2002) 277:19289–19294. https://doi.org/10.1074/jbc.M201572200 [DOI] [PubMed] [Google Scholar]
- [68].Bridges R, Lutgen V, Lobner D, Baker DA, Thinking outside the cleft to understand synaptic activity: contribution of the cystine-glutamate antiporter (System xc-) to normal and pathological glutamatergic signaling. Pharmacological Reviews. (2012) 64: 780–802. 10.1124/pr.110.003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ghosh HS, McBurney M, Robbins PD, SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. (2010) 5(2):e9199. 10.1371/journal.pone.0009199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liu Y, Wu Y, Diao Z, Guo W, Liu W, Resveratrol inhibits parathyroid hormone-induced apoptosis in human aortic smooth muscle cells by upregulating sirtuin 1. Ren Fail. (2019b) 41:401–407. 10.1080/0886022X.2019.1605296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wang X, Zhang Y, Resveratrol alleviates LPS-induced injury in human keratinocyte cell line HaCaT by up-regulation of miR-17. Biochem Biophys Res Commun. (2018)501:106–112. 10.1016/j.bbrc.2018.04.184 [DOI] [PubMed] [Google Scholar]
- [72].Kang Y, Zhang H, Zhao Y, Wang Y, Wang W, He Y, Zhang W, Zhang W, Zhu X, Zhou Y, Zhang L, Ju Z, Shi L, Telomere Dysfunction Disturbs Macrophage Mitochondrial Metabolism and the NLRP3 Inflammasome through the PGC-1α/TNFAIP3 Axis. Cell Rep. (2018) 22:3493–3506. 10.1016/j.celrep.2018.02.071 [DOI] [PubMed] [Google Scholar]
- [73].Mattson MP, Barger SW, Furukawa K, Bruce AJ, Wyss-Coray T, Mark RJ, Mucke L, Cellular signaling roles of TGF beta, TNF alpha and beta APP in brain injury responses and Alzheimer’s disease. Brain Res Brain Res Rev. (1997) 23:47–61. 10.1016/S0165-0173(96)00014-8 [DOI] [PubMed] [Google Scholar]
- [74].Hafner A, Bulyk ML, Jambhekar A, Lahav G, The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. (2019) 20:199–210. 10.1038/s41580-019-0110-x [DOI] [PubMed] [Google Scholar]
- [75].McCubrey JA, Lertpiriyapong K, Fitzgerald TL, Martelli AM, Cocco L, Rakus D, Gizak A, Libra M, Cervello M, Montalto G, Yang LV, Abrams SL, Steelman LS, Roles of TP53 in determining therapeutic sensitivity, growth, cellular senescence, invasion and metastasis. Adv Biol Regul. (2017) 63:32–48. 10.1016/j.jbior.2016.10.001. [DOI] [PubMed] [Google Scholar]
- [76].Rufini A, Tucci P, Celardo I, Melino G, Senescence and aging: the critical roles of p53. Oncogene. (2013) 32:5129–5143. 10.1038/onc.2012.640 [DOI] [PubMed] [Google Scholar]
- [77].Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Park SH, Thompson T, Karsenty G, Bradley A, Donehower LA, p53 mutant mice that display early ageing-associated phenotypes. Nature. (2002) 415(6867): 45–53. 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- [78].Negis Y, Karabay A, Expression of cell cycle proteins in cortical neurons-Correlation with glutamate-induced neurotoxicity. Biofactors. (2016) 42:358–367. 10.1002/biof.1282 [DOI] [PubMed] [Google Scholar]
- [79].Vogler M, BCL2A1: the underdog in the BCL2 family. Cell Death Differ. (2012)19:67–74. 10.1038/cdd.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bellaver B, Souza DG, Souza DO, Quincozes-Santos A, Hippocampal Astrocyte Cultures from Adult and Aged Rats Reproduce Changes in Glial Functionality Observed in the Aging Brain. Mol Neurobiol. (2017) 54:2969–2985. 10.1007/s12035-016-9880-8 [DOI] [PubMed] [Google Scholar]
- [81].Childs Bennett G, Durik Matej, Baker Darren J, van Deursen Jan M, Cellular senescence in aging and age-related disease: from mechanisms to therapy Nat Med. (2015) 21:1424–1435. 10.1038/nm.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Diep CH, Knutson TP, Lange CA, Active FOXO1 Is a Key Determinant of Isoform-Specific Progesterone Receptor Transactivation and Senescence Programming. Mol Cancer Res. (2016) 14:141–162. 10.1158/1541-7786.MCR-15-0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Matthaei M, Meng H, Meeker AK, Eberhart CG, Jun AS, Endothelial Cdkn1a (p21) overexpression and accelerated senescence in a mouse model of Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. (2012) 53:6718–6727. https://www.ncbi.nlm.nih.gov/pubmed/22956607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Patil M, Pabla N, Dong Z, Checkpoint kinase 1 in DNA damage responseand cell cycle regulation”. Cellular and Molecular Life Sciences. (2013) 70: 4009–4021. https://doi.org/10.1007/s00018-013-1307-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Edwards MG, Anderson RM, Yuan M, Kendziorski CM, Weindruch R, Prolla TA, Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. (2007)8:80 10.1186/1471-2164-8-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kletsas D, Pratsinis H, Mariatos G, Zacharatos P, Gorgoulis VG, The proinflammatory phenotype of senescent cells: the p53-mediated ICAM-1 expression. Ann N Y Acad Sci. (2004)1019:330–332. 10.1196/annals.1297.056 [DOI] [PubMed] [Google Scholar]
- [87].de Oliveira LR, Mombach JC, Castellani G, A simple stochastic model for the feedback circuit between p16INK4a and p53 mediated by p38MAPK: implications for senescence and apoptosis. Mol Biosyst. (2015) 11:2955–2963. 10.1039/c5mb00230c [DOI] [PubMed] [Google Scholar]
- [88].Xiao P, Huang X, Huang L, Yang J, Li A, Shen K, Wedegaertner PB, Jiang X, G protein-coupled receptor kinase 4-induced cellular senescence and its senescence-associated gene expression profiling. Exp Cell Res. (2017) 360:273–280. 10.1016/j.yexcr.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gao Q, Chen K, Gao L, Zheng Y, Yang YG, Thrombospondin-1 signaling through CD47 inhibits cell cycle progression and induces senescence in endothelial cells. Cell Death Dis. (2016) 7:e2368. 10.1038/cddis.2016.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].LeBlanc AJ, Kelm NQ, Thrombospondin-1, Free Radicals, and the Coronary Microcirculation: The Aging Conundrum. Antioxid Redox Signal. (2017) 27:785–801. 10.1089/ars.2017.7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Meijles DN, Sahoo S, Al Ghouleh I, Amaral JH, Bienes-Martinez R, Knupp HE, Attaran S, Sembrat JC, Nouraie SM, Rojas MM, Novelli EM Gladwin MT, Isenberg JS, Cifuentes-Pagano E, Pagano PJ, The matricellular protein TSP1 promotes human and mouse endothelial cell senescence through CD47 and Nox1. Sci Signal. (2017) 10(501). pii: eaaj1784. https://doi.org/10.1126/scisignal.aaj1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yue X, Wu L, Hu W, The regulation of leukemia inhibitory factor. Cancer Cell Microenviron. (2015)2 pii: e877. https://doi.org/10.14800/ccm.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hänzelmann S, Beier F, Gusmao EG, Koch CM, Hummel S, Charapitsa I, Joussen S, Benes V, Brümmendorf TH, Reid G, Costa IG, Wagner W, Replicative senescence is associated with nuclear reorganization and with DNA methylation at specific transcription factor binding sites. Clin Epigenetics. (2015) 7:19 10.1186/s13148-015-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].McPherson LA, Loktev AV, Weigel RJ, Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. (2002) 277:45028–45033. 10.1074/jbc.M208924200 [DOI] [PubMed] [Google Scholar]
- [95].Bennett RE, Robbins AB, Hu M, Cao X, Betensky RA, Clark T, Das S, Hyman BT, Tau induces blood vessel abnormalities and angiogenesis-related gene expression in P301L transgenic mice and human Alzheimer’s disease. Proc Natl Acad Sci U S A. (2018) 115:E1289–E1298. https://doi.org/10.1073/pnas.1710329115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Khan SS, Shah SJ, Klyachko E, Baldridge AS, Eren M, Place AT, Aviv A, Puterman E, Lloyd-Jones DM, Heiman M, Miyata T, Gupta S, Shapiro AD, Vaughan DE, A null mutation in SERPINE1 protects against biological aging in humans. Sci Adv. (2017) 3(11):eaao1617. 10.1126/sciadv.aao1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tsikouris JP, Cox CD, Pharmacologic blockade of the renin-angiotensin system: vascular benefits beyond commonly understood pharmacologic actions. Pharmacotherapy. (2003) 23:1141–1152. 10.1177/009127002762491271 [DOI] [PubMed] [Google Scholar]
- [98].Fogari R, Zoppi A, Mugellini A, Maffioli P, Lazzari P, Derosa G, Role of angiotensin II in plasma PAI-1 changes induced by imidapril or candesartan in hypertensive patients with metabolic syndrome. Hypertens Res. (2011) 34:1321–1326. 10.1038/hr.2011.137 [DOI] [PubMed] [Google Scholar]
- [99].Boe AE, Eren M, Murphy SB, Kamide CE, Ichimura A, Terry D, McAnally D, Smith LH, Miyata T, Vaughan DE, Plasminogen activator inhibitor-1 antagonist TM5441 attenuates Nω-nitro-L-arginine methyl ester-induced hypertension and vascular senescence. Circulation. (2013) 128:2318–2324. 10.1161/CIRCULATIONAHA.113.003192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gorska-Ciebiada M, Saryusz-Wolska M, Borkowska A, Ciebiada M, Loba J, Plasma levels of thrombomodulin, plasminogen activator inhibitor-1 and fibrinogen in elderly, diabetic patients with depressive symptoms. Aging Clin Exp Res. (2016) 28:843–851. 10.1007/s40520-015-0504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Miskin R, Masos T, Yahav S, Shinder D, Globerson A, AlphaMUPA mice: a transgenic model for increased life span. Neurobiol Aging. (1999) 20:555–564. PMID:10638529 [DOI] [PubMed] [Google Scholar]
- [102].Savoy C, Van Lieshout RJ, Steiner M, Is plasminogen activator inhibitor-1 a physiological bottleneck bridging major depressive disorder and cardiovascular disease? Acta Physiol (Oxf). (2017) 219:715–727. 10.1111/apha.12726 [DOI] [PubMed] [Google Scholar]
- [103].Vaughan DE, Rai R, Khan SS, Eren M, Ghosh AK, Plasminogen Activator Inhibitor-1 Is a Marker and a Mediator of Senescence. Arterioscler Thromb Vasc Biol. (2017) 37:1446–1452. 10.1161/ATVBAHA.117.309451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL, Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest. (2013)123:966–972. 10.1172/JCI64098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Pawlak R, Magarinos AM, Melchor J, McEwen B, Strickland S, Tissue plasminogen activator in the amygdala is critical for stress-induced anxiety-like behavior. Nat Neurosci. (2003) 6:168–174. 10.1038/nn998 [DOI] [PubMed] [Google Scholar]
- [106].Zhang T, Tian F, Wang J, Zhou S, Dong X, Guo K, Jing J, Zhou Y, Chen Y, Donepezil attenuates high glucose-accelerated senescence in human umbilical vein endothelial cells through SIRT1 activation. Cell Stress Chaperones. (2015)20:787–792. 10.1007/s12192-015-0601-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zou H, Stoppani E, Volonte D, Galbiati F, Caveolin-1, cellular senescence and age-related diseases. Mech Ageing Dev. (2011)132:533–542. https://doi.org/10.1016/j.mad.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bose S, Cho J, Role of chemokine CCL2 and its receptor CCR2 in neurodegenerative diseases. Arch Pharm Res. (2013) 36:1039–1050. 10.1007/s12272-013-0161-z [DOI] [PubMed] [Google Scholar]
- [109].Li Y, Ohms SJ, Sun C, Fan J, NF-κB controls Il2 and Csf2 expression during T cell development and activation process. Mol Biol Rep. (2013) 40:1685–1692. https://doi.org/10.1007/s11033-012-2219-2 [DOI] [PubMed] [Google Scholar]
- [110].Imai R, Asai K, Hanai J, Takenaka M, Transgenic mice overexpressing glia maturation factor-β, an oxidative stress inducible gene, show premature aging due to Zmpste24 down-regulation. Aging (Albany NY). (2015) 7:486–499. https://doi.org/10.18632/aging.100779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Seo Y, Kim HS, Kang I, Choi SW, Shin TH, Shin JH, Lee BC, Lee JY, Kim JJ, Kook MG, Kang KS, Cathepsin S contributes to microglia-mediated olfactory dysfunction through the regulation of Cx3cl1-Cx3cr1 axis in a Niemann-Pick disease type C1 model. Glia. (2016) 64:2291–2305. 10.1002/glia.23077 [DOI] [PubMed] [Google Scholar]
- [112].Pišlar AH, Zidar N, Kikelj D, Kos J, Cathepsin X promotes 6-hydroxydopamine-induced apoptosis of PC12 and SH-SY5Y cells. Neuropharmacology. (2014) 82:121–131. 10.1016/j.neuropharm.2013.07.040 [DOI] [PubMed] [Google Scholar]
- [113].Liu X, Fu B, Chen D, Hong Q, Cui J, Li J, Bai X, Chen X, miR-184 and miR-150 promote renal glomerular mesangial cell aging by targeting Rab1a and Rab31. Exp Cell Res. (2015) 336:192–203. 10.1016/j.yexcr.2015.07.006 [DOI] [PubMed] [Google Scholar]
- [114].Castillo E, Leon J, Mazzei G, Abolhassani N, Haruyama N, Saito T, Saido T, Hokama M, Iwaki T, Ohara T, Ninomiya T, Kiyohara Y, Sakumi K, LaFerla FM, Nakabeppu Y, Comparative profiling of cortical gene expression in Alzheimer’s disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep. (2017) 7:17762 10.1038/s41598-017-17999-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Khemais-Benkhiat S, Belcastro E, Idris-Khodja N, Park SH, Amoura L, Abbas M, Auger C, Kessler L, Mayoux E, Toti F, Schini-Kerth VB, Angiotensin II-induced redox-sensitive SGLT1 and 2 expression promotes high glucose-induced endothelial cell senescence. J Cell Mol Med. (2019) March 30 10.1111/jcmm.14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Rajagopalan S, Brook R, Mehta RH, Supiano M, Pitt B, Effect of losartan in aging-related endothelial impairment. Am J Cardiol. (2002) 89:562–566. 10.1016/S0002-9149(01)02297-4 [DOI] [PubMed] [Google Scholar]
- [117].Buga AM, Margaritescu C, Scholz CJ, Radu E, Zelenak C, Popa-Wagner A, Transcriptomics of post-stroke angiogenesis in the aged brain. Front Aging Neurosci. (2014) 6:44 10.3389/fnagi.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shen ZJ, Xu CS, Li YP, Li J, Xu JJ, Xia P, Telmisartan inhibits Ang II-induced MMP-9 expression in macrophages in stabilizing atheromatous plaque. Eur Rev Med Pharmacol Sci. (2018) 22:8004–8012. 10.26355/eurrev_201811_16429 [DOI] [PubMed] [Google Scholar]
- [119].Rodier F, Coppé JP, Patil CK, Hoeijmakers WA, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J, Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. (2009) 11:973–979. 10.1038/ncb1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Andriani GA, Almeida VP, Faggioli F, Mauro M, Tsai WL, Santambrogio L, Maslov A, Gadina M, Campisi J, Vijg J, Montagna C, Whole Chromosome Instability induces senescence and promotes SASP. Sci Rep. (2016) 6:35218 10.1038/srep35218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Fernández-Duran I, Tarrats N, Hari P, Acosta JC, Measuring the Inflammasome in Oncogene-Induced Senescence. Methods Mol Biol. (2019) 1896:57–70. 10.1007/978-1-4939-8931-7_7 [DOI] [PubMed] [Google Scholar]
- [122].Franceschi C, Campisi J, Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. (2014) 69 Suppl 1:S4–9. https://doi.org/10.1093/gerona/glu057 [DOI] [PubMed] [Google Scholar]
- [123].Michaud M, Balardy L, Moulis G, Gaudin C, Peyrot C, Vellas B, Cesari M, Nourhashemi F, Proinflammatory cytokines, aging, and age-related diseases. J Am Med Dir Assoc. (2013) 14:877–882. 10.1016/j.jamda.2013.05.009 [DOI] [PubMed] [Google Scholar]
- [124].Dominick G, Bowman J, Li X, Miller RA, Garcia GG, mTOR regulates the expression of DNA damage response enzymes in long-lived Snell dwarf, GHRKO, and PAPPA-KO mice. Aging Cell. (2017)16: 52–60. 10.1111/acel.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Fang BA, Kovačević Ž, Park KC, Kalinowski DS, Jansson PJ, Lane DJ, Sahni S, Richardson DR, Molecular functions of the iron-regulated metastasis suppressor, NDRG1, and its potential as a molecular target for cancer therapy. Biochim. Biophys. Acta. (2014) 1845: 1–19. https://doi.org/10.1016/j.bbcan.2013.11.002 [DOI] [PubMed] [Google Scholar]
- [126].Chung KW, Jeong HO, Lee B, Park D, Kim DH, Choi YJ, Lee EK, Kim KM, Park JW, Yu BP, Chung HY, Involvement of NF-κBIZ and related cytokines in age-associated renal fibrosis. Oncotarget. (2017) 8:7315–7327. 10.18632/oncotarget.14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Lim H, Park H, Kim HP. Effects of flavonoids on senescence-associated secretory phenotype formation from bleomycin-induced senescence in BJ fibroblasts. Biochem Pharmacol. 2015. August 15;96(4):337–48. 10.1016/j.bcp.2015.06.013 [DOI] [PubMed] [Google Scholar]
- [128].Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Jänicke RU, Schulze-Osthoff K, A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem. (2006) 281:12645–12654. 10.1074/jbc.M511956200 [DOI] [PubMed] [Google Scholar]
- [129].Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, Saitoh T, Yamaoka S, Yamamoto N, Yamamoto S, Muta T, Takeda K, Akira S, Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. (2004)430: 218–222. 10.1038/nature02738 [DOI] [PubMed] [Google Scholar]
- [130].Özcan S, Alessio N, Acar MB, Mert E, Omerli F, Peluso G, Galderisi U, Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging (Albany NY). (2016) 8:1316–1329. 10.18632/aging.100971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Danka Mohammed CP, Park JS, Nam HG, Kim K, MicroRNAs in brain aging. Mech Ageing Dev. (2017)168:3–9. 10.1016/j.mad.2017.01.007 [DOI] [PubMed] [Google Scholar]
- [132].DiLoreto R, Murphy CT, The cell biology of aging. Mol Biol Cell. (2015) 26:4524–4531. 10.1091/mbc.E14-06-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Soto-Gamez A, Demaria M, Therapeutic interventions for aging: the case of cellular senescence. Drug Discov Today. (2017) 22:786–795. 10.1016/j.drudis.2017.01.004 [DOI] [PubMed] [Google Scholar]
- [134].Hammond SM, Sharpless NE, HMGA2, microRNAs, and stem cell aging. Cell. (2008) 135:1013–1016. 10.1016/j.cell.2008.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].D’Souza T, Sherman-Baust CA, Poosala S, Mullin JM, Morin PJ, Age-related changes of claudin expression in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med Sci. (2009) 64:1146–1153. 10.1093/gerona/gp118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Petit MM, Meulemans SM, Van de Ven WJ, The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J Biol Chem. (2003) 278:2157–2168. 10.1074/jbc.M206106200 [DOI] [PubMed] [Google Scholar]
- [137].Hinman JD, Chen CD, Oh SY, Hollander W, Abraham CR, Age-dependent accumulation of ubiquitinated 2’,3’-cyclic nucleotide 3’-phosphodiesterase in myelin lipid rafts. Glia. (2008) 56: 118–133. 10.1002/glia.20595 [DOI] [PubMed] [Google Scholar]
- [138].Krestinina O, Azarashvili T, Baburina Y, Galvita A, Grachev D, Stricker R, Reiser G, In aging, the vulnerability of rat brain mitochondria is enhanced due to reduced level of 2’,3’-cyclic nucleotide-3’-phosphodiesterase (CNP) and subsequently increased permeability transition in brain mitochondria in old animals. Neurochem Int. (2015) 80:41–50. 10.1016/j.neuint.2014.09.008 [DOI] [PubMed] [Google Scholar]
- [139].Campos-Chillon F, Farmerie TA, Bouma GJ, Clay CM, Carnevale EM, Effects of aging on gene expression and mitochondrial DNA in the equine oocyte and follicle cells. Reprod Fertil Dev. (2015) 27:925–933. 10.1071/RD14472 [DOI] [PubMed] [Google Scholar]
- [140].Fujita Y, Taniguchi Y, Shinkai S, Tanaka M, Ito M, Secreted growth differentiation factor 15 as a potential biomarker for mitochondrial dysfunctions in aging and age-related disorders. Geriatr Gerontol Int. (2016) 16 Suppl 1:17–29. 10.1111/ggi.12724 [DOI] [PubMed] [Google Scholar]
- [141].Park H, Kim CH, Jeong JH, Park M, Kim KS, GDF15 contributes to radiation-induced senescence through the ROS-mediated p16 pathway in human endothelial cells. Oncotarget. (2016) 7:9634–9644. 10.18632/oncotarget.7457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Agrawal A, Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: a mini-review. Gerontology (2013) 59:421–426. https://doi.org/10.1159/000350536 [DOI] [PubMed] [Google Scholar]
- [143].Cheriyath V, Leaman DW, Borden EC, Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. (2011) 31:173–181. 10.1089/jir.2010.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Bortell N, Basova L, Najera JA, Morsey B, Fox HS, Marcondes MCG, Sirtuin 1-Chromatin-Binding Dynamics Points to a Common Mechanism Regulating Inflammatory Targets in SIV Infection and in the Aging Brain. J Neuroimmune Pharmacol. (2018)13:163–178. 10.1007/s11481-017-9772-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Dhib-Jalbut S, The choroid plexus and the paradox of interferons in the aging brain. Cytokine. (2015) 71:413–414. 10.1016/j.cyto.2014.11.021 [DOI] [PubMed] [Google Scholar]
- [146].Li Q, Tang L, Roberts PC, Kraniak JM, Fridman AL, Kulaeva OI, Tehrani OS, Tainsky MA, Interferon regulatory factors IRF5 and IRF7 inhibit growth and induce senescence in immortal Li-Fraumeni fibroblasts. Mol Cancer Res. (2008) 6:770–784. 10.1158/1541-7786.MCR-07-0114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Ning S, Pagano JS, Barber GN, IRF7: activation, regulation, modification and function. Genes Immun. (2011)12:399–414. https://doi.org/10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Rodriguez-Miguelez P, Fernandez-Gonzalo R, Almar M, Mejías Y, Rivas A, de Paz JA, Cuevas MJ, González-Gallego J, Role of Toll-like receptor 2 and 4 signaling pathways on the inflammatory response to resistance training in elderly subjects. Age (Dordr). (2014)36:9734 10.1007/s11357-014-9734-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Klosinski LP, Yao J, Yin F, Fonteh AN, Harrington MG, Christensen TA, Trushina E, Brinton RD, White Matter Lipids as a Ketogenic Fuel Supply in Aging Female Brain: Implications for Alzheimer’s Disease. EBioMedicine. (2015) 2:1888–1904. 10.1016/j.ebiom.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tang B, Capitao C, Dean B, Thomas EA, Differential age-and disease-related effects on the expression of genes related to the arachidonic acid signaling pathway in schizophrenia. Psychiatry Res. (2012)196:201–206. 10.1016/j.psychres.2011.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kim J, Vaish V, Feng M, Field K, Chatzistamou I, Shim M, Transgenic expression of cyclooxygenase-2 (COX2) causes premature aging phenotypes in mice. Aging (Albany NY). (2016) 8:2392–2406. 10.18632/aging.101060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Kang MK, Kameta A, Shin KH, Baluda MA, Kim HR, Park NH, Senescence-associated genes in normal human oral keratinocytes. Exp Cell Res. (2003) 15;287:272–281. 10.1016/S0014-4827(03)00061-2 [DOI] [PubMed] [Google Scholar]
- [153].White CC, Yang HS, Yu L, Chibnik LB, Dawe RJ, Yang J, Klein HU, Felsky D, Ramos-Miguel A, Arfanakis K, Honer WG, Sperling RA, Schneider JA, Bennett DA, De Jager PL, Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med. (2017)14:e1002287. 10.1371/journal.pmed.1002287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Iram T, Trudler D, Kain D, Kanner S, Galron R, Vassar R, Barzilai A, Blinder P, Fishelson Z, Frenkel D, Astrocytes from old Alzheimer’s disease mice are impaired in Aβ uptake and in neuroprotection. Neurobiol Dis. (2016) 96:84–94. 10.1016/j.nbd.2016.08.001 [DOI] [PubMed] [Google Scholar]
- [155].Hossain MN, Sakemura R, Fujii M, Ayusawa D, G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun. (2006)351:645–650. 10.1016/j.bbrc.2006.10.112 [DOI] [PubMed] [Google Scholar]
- [156].Nelson PT, Wang WX, Partch AB, Monsell SE, Valladares O, Ellingson SR, Wilfred BR, Naj AC, Wang LS, Kukull WA, Fardo DW, Reassessment of risk genotypes (GRN, TMEM106B, and ABCC9 variants) associated with hippocampal sclerosis of aging pathology. J Neuropathol Exp Neurol. (2015) 74:75–84. 10.1097/NEN.0000000000000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Pienimaeki-Roemer A, Konovalova T, Musri MM, Sigruener A, Boettcher A, Meister G, Schmitz G, Transcriptomic profiling of platelet senescence and platelet extracellular vesicles. Transfusion. (2017) 57:144–156. 10.1111/trf.13896 [DOI] [PubMed] [Google Scholar]
- 158.[] Petyuk VA, Chang R, Ramirez-Restrepo M, Beckmann ND, Henrion MYR, Piehowski PD, Zhu K, Wang S, Clarke J, Huentelman J, Xie F, Andreev V, Engel A, Guettoche T, Navarro L, De Jager P, Schneider JA, Morris CM, McKeith IG, Perry RH, Lovestone S, Woltjer RL, Beach TG, Sue LI, Serrano GE, Lieberman AP, Albin RL, Ferrer I, Mash DC, Hulette CM, Ervin JF, Reiman EM, Hardy JA, Bennett DA, Schadt E, Smith RD, Myers AJ, The human brainome: network analysis identifies HSPA2 as a novel Alzheimer’s disease target. Brain. (2018) 141:2721–2739. 10.1093/brain/awy215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Cardoso AL, Fernandes A, Aguilar-Pimentel JA, de Angelis MH, Guedes JR, Brito MA, Ortolano S, Pani G, Athanasopoulou S, Gonos ES, Schosserer M, Grillari J, Peterson P, Tuna BG, Dogan S, Meyer A, van Os R, Trendelenburg AU, Towards frailty biomarkers: Candidates from genes and pathways regulated in aging and age-related diseases. Ageing Res Rev. (2018) 214–277. 10.1016/j.arr.2018.07.004 [DOI] [PubMed] [Google Scholar]
- [160].Dietz K, de Los Reyes Jiménez M, Gollwitzer ES, Chaker AM, Zissler UM, Rådmark OP, Baarsma HA, Königshoff M, Schmidt-Weber CB, Marsland BJ, Esser-von Bieren J, Age dictates a steroid-resistant cascade of Wnt5a, transglutaminase 2, and leukotrienes in inflamed airways. J Allergy Clin Immunol. (2017)139:1343–1354.e6. 10.1016/j.jaci.2016.07.014 [DOI] [PubMed] [Google Scholar]
- [161].Ricotta M, Iannuzzi M, Vivo GD, Gentile V, Physio-pathological roles of transglutaminase-catalyzed reactions. World Journal of Biological Chemistry. (2010) 1: 181–187. 10.4331/wjbc.v1.i5.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Wilhelmus MM, Verhaar R, Andringa G, Bol JG, Cras P, Shan L, Hoozemans JJ, Drukarch B, Presence of tissue transglutaminase in granular endoplasmic reticulum is characteristic of melanized neurons in Parkinson’s disease brain. Brain Pathology.(2011) 21:130–139. 10.1111/j.1750-3639.2010.00429.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Hsiao R, Greenberg B, Neprilysin Inhibition as a PARADIGM Shift in Heart Failure Therapy. Curr Heart Fail Rep. (2016) 13:172–180. 10.1007/s11897-016-0297-5 [DOI] [PubMed] [Google Scholar]
- [164].Liu XL, Liu JL, Xu YC, Zhang X, Wang YX, Qing LH, Guo W, Ding J, Meng LH, Membrane metallo-endopeptidase mediates cellular senescence induced by oncogenic PIK3CAH1047R accompanied with pro-tumorigenic secretome. Int J Cancer. (2019a) 145:817–829. 10.1002/ijc.32153 [DOI] [PubMed] [Google Scholar]
- [165].Benton MC, Sutherland HG, Macartney-Coxson D, Haupt LM, Lea RA, Griffiths LR, Methylome-wide association study of whole blood DNA in the Norfolk Island isolate identifies robust loci associated with age. Aging (Albany NY). (2017) 9:753–768. 10.18632/aging.101187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Yu L, Petyuk VA, Gaiteri C, Mostafavi S, Young-Pearse T, Shah RC, Buchman AS, Schneider JA, Piehowski PD, Sontag RL, Fillmore TL, Shi T, Smith RD, De Jager PL, Bennett DA, Targeted brain proteomics uncover multiple pathways to Alzheimer’s dementia. Ann Neurol. (2018) 84:78–88. https://doi.org/10.1002/ana.25266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Zhang H, Jia Y, Cooper JJ, Hale T, Zhang Z, Elbein SC, Common variants in glutamine:fructose-6-phosphate amidotransferase 2 (GFPT2) gene are associated with type 2diabetes, diabetic nephropathy, and increased GFPT2 mRNA levels. J Clin Endocrinol Metab. (2004)89:748–755. 10.1210/jc.2003-031286 [DOI] [PubMed] [Google Scholar]
- [168].Kirby TJ, Walton RG, Finlin B, Zhu B, Unal R, Rasouli N, Peterson CA, Kern PA, Integrative mRNA-microRNA analyses reveal novel interactions related to insulin sensitivity in human adipose tissue. Physiol Genomics. (2016) 48:145–153. 10.1152/physiolgenomics.00071.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Van Kirk CA, VanGuilder HD, Young M, Farley JA, Sonntag WE, Freeman WM, Age-related alterations in retinal neurovascular and inflammatory transcripts. Mol Vis. (2011)17:1261–1274. PMID: 21633715 [PMC free article] [PubMed] [Google Scholar]
- [170].Verdier V, Csárdi G, de Preux-Charles AS, Médard JJ, Smit AB, Verheijen MH, Bergmann S, Chrast R, Aging of myelinating glial cells predominantly affects lipid metabolism and immune response pathways. Glia. (2012) 60:751–760. 10.1002/glia.22305 [DOI] [PubMed] [Google Scholar]
- [171].Kojima T, Kamei H, Aizu T, Arai Y, Takayama M, Nakazawa S, Ebihara Y, Inagaki H, Masui Y, Gondo Y, Sakaki Y, Hirose N, Association analysis between longevity in the Japanese population and polymorphic variants of genes involved in insulin and insulin-like growth factor 1 signaling pathways. Exp Gerontol. (2004) 39:1595–1598. 10.1016/j.exger.2004.05.007 [DOI] [PubMed] [Google Scholar]
- [172].Oudit GY, Kassiri Z, Patel MP, Chappell M, Butany J, Backx PH, Tsushima RG, Scholey JW, Khokha R, Penninger JM, Angiotensin II-mediated oxidative stress and inflammation mediate the age-dependent cardiomyopathy in ACE2 null mice. Cardiovasc Res. (2007) 75:29–39. 10.1016/j.cardiores.2007.04.007 [DOI] [PubMed] [Google Scholar]
- [173].Qiu L, Yu H, Liang F, Multiple C2 domains transmembrane protein 1 is expressed in CNS neurons and possibly regulates cellular vesicle retrieval and oxidative stress. J Neurochem. (2015) 135:492–507. 10.1111/jnc.13251 [DOI] [PubMed] [Google Scholar]
- [174].Shakeri H, Gevaert AB, Schrijvers DM, De Meyer GRY, De Keulenaer GW, Guns PDF, Lemmens K, Segers VF, Neuregulin-1 attenuates stress-induced vascular senescence. Cardiovasc Res. (2018)114:1041–1051. 10.1093/cvr/cvy059 [DOI] [PubMed] [Google Scholar]
- [175].Lenkowski JR, Qin Z, Sifuentes CJ, Thummel R, Soto CM, Moens CB, Raymond PA, Retinal regeneration in adult zebrafish requires regulation of TGFβ signaling. Glia. (2013) 61:1687–1697. https://doi.org/10.1002/glia.22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Zerlanko BJ, Bartholin L, Melhuish TA, Wotton D, Premature senescence and increased TGFβ signaling in the absence of Tgif1. PLoS One. (2012)7:e35460. 10.1371/journal.pone.0035460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Bernard JA, Seidler RD. Moving forward: age effects on the cerebellum underlie cognitive and motor declines. Neurosci Biobehav Rev. (2014) 42:193–207. 10.1016/j.neubiorev.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Canu N, Calissano P. In vitro cultured neurons for molecular studies correlating apoptosis with events related to Alzheimer disease. Cerebellum. (2003) 2:270–278. 10.1080/14734220310004289 [DOI] [PubMed] [Google Scholar]
- [179].Contestabile A Cerebellar granule cells as a model to study mechanisms of neuronal apoptosis or survival in vivo and in vitro. Cerebellum. (2002) 1:41–55. 10.1080/147342202753203087 [DOI] [PubMed] [Google Scholar]
- [180].Hoxha E, Lippiello P, Zurlo F, Balbo I, Santamaria R, Tempia F, Miniaci MC. The Emerging Role of Altered Cerebellar Synaptic Processing in Alzheimer’s Disease. Front Aging Neurosci. (2018) November 27;10:396 10.3389/fnagi.2018.00396.eCollection2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Lackey EP, Heck DH, Sillitoe RV. Recent advances in understanding the mechanisms of cerebellar granule cell development and function and their contribution to behavior. F1000Res. (2018) July 26;7 pii: F1000 Faculty Rev-1142. 10.12688/f1000research.15021.1eCollection2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Mavroudis I Cerebellar pathology in Alzheimer’s disease. Hell J Nucl Med. (2019) Jan-Apr;22 Suppl:174–179. PMID: 30877735 [PubMed] [Google Scholar]
- [183].Radio N, Frank S. Neuronal Cell Morphology in Primary Cerebellar Granule Cells Using High-Content Analysis. Methods Mol Biol. (2018) 1727:227–237. http: 10.1007/978-1-4939-7571-6_17 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole Gene Set Enrichment Analysis output for all aging associated genesets with plots, statistics, gene status and heatmaps. GO: Gene Ontology Data Archive. Genesets with links and References are listed in Table 1. :
The table shows the whole Gene Set Enrichment Analysis output for all senescence associated genesets with plots, statistics, gene status and heatmaps. GO: Gene Ontology Data Archive. Genesets with links and References are listed in Table 1.
We looked for age upregulated genes in 6 different brain aging datasets (GSE11882_PCG, GSE11882_SFG, GSE11882_Hippo, GSE11882_EnthorinalCrtx, GSE11697_HippoCA1 and GSE17757_SFG). 116 genes were consistently up-regulated by 1.2 folds in all 6 different brain tissues aging datasets (blue columns) and 99 of them are also present in the CGC dataset (Green columns). Of the 99 genes upregulated in aging and present in the CGC study, 70 of them are also upregulated by Glutamate alone and are down-regulated or normalized by Candesartan treatment. In each comparison dataset, the p-value, Fold Change and change direction are represented. Aging Data are taken from GSE11882, GSE11697 and GSE17757 (see detailed information in Table 1). GoRilla analysis is presented in this order: GoRilla_process GoRilla_Function, GoRilla_Component (description, p-value, FDR q-value, enrichment, and genes), GO_BP, GO_MF, GO_CC, GO_diseases, GO_GAD_Disease (category, term, RT, genes, count, %, p-value, Benjamini).
The list of all the IPA up-stream regulators for the aging associated geneset (grey rows, 1101 regulators) and the CGC glutamate up-regulated and Candesartan down-regulated geneset (yellow rows, 1203 regulators) were sorted based on the upstream regulator. The regulator that are unique to the CGC glutamate+Candesartan are labeled in red and the ones unique to the aging dataset are in blue. The table includes their predicted activation state, the activation z-score, the p-value of overlap, the target molecules and whether the upstream regulators are common or unique, referring to regulators that are common in both genesets or unique to aging (blue label) or unique to Glutamate/Candesartan (red label). The negative z-score is associated with predicted inhibition of the upstream regulator and the positive z-score is associated with predictive activation of the upstream regulator (Combined upstream regulators). A list of 28 representative upstream regulators based on known and published genes, compounds and pathways associated with aging includes z-scores and p-values for GSE11882_Aging_UP and for CGC_Glutamate-UP_Candesartan-Down that are upregulated consistently in 6 different aging datasets (Table 2).
List of genes that are GSEA enriched in at least 3 out of 19 aging and senescence datasets from different tissues and cells studied.