Figure 1 |. Overview of non-homologous end joining.
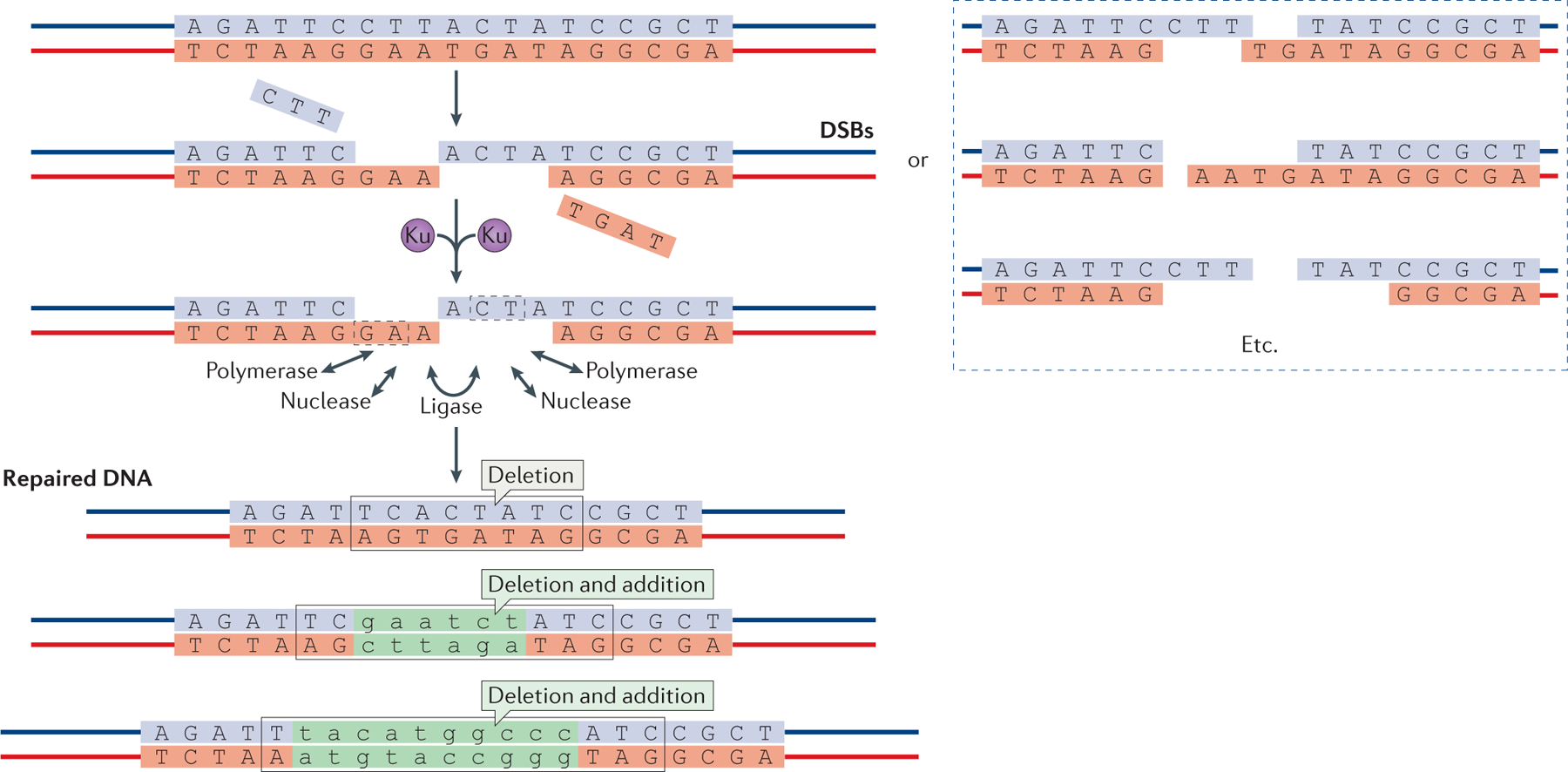
Schematic of DNA double-strand breaks (DSBs) and their repair by non-homologous end joining (NHEJ) (top). The Ku70–Ku80 heterodimer binds to DSBs and improves their subsequent binding by the NHEJ polymerase, nuclease and ligase complexes. These enzymes can act on DSBs in any order to resect and add nucleotides. Multiple rounds of resection and addition are possible, and nuclease and polymerase activities at each of the two DNA ends seem to be independent. Microhomology between the two DNA ends, which is either already present (dashed boxes) or newly created when the polymerases add nucleotides in a template-independent manner, is often used to guide end joining. The process is error-prone and can result in diverse DNA sequences at the repair junction (bottom). However, NHEJ is also capable of joining two DNA ends without nucleotide loss from either DNA end and without any addition. Nucleotide additions are depicted in green lower case.
