Figure 3 |. The various non-homologous end joining subpathways.
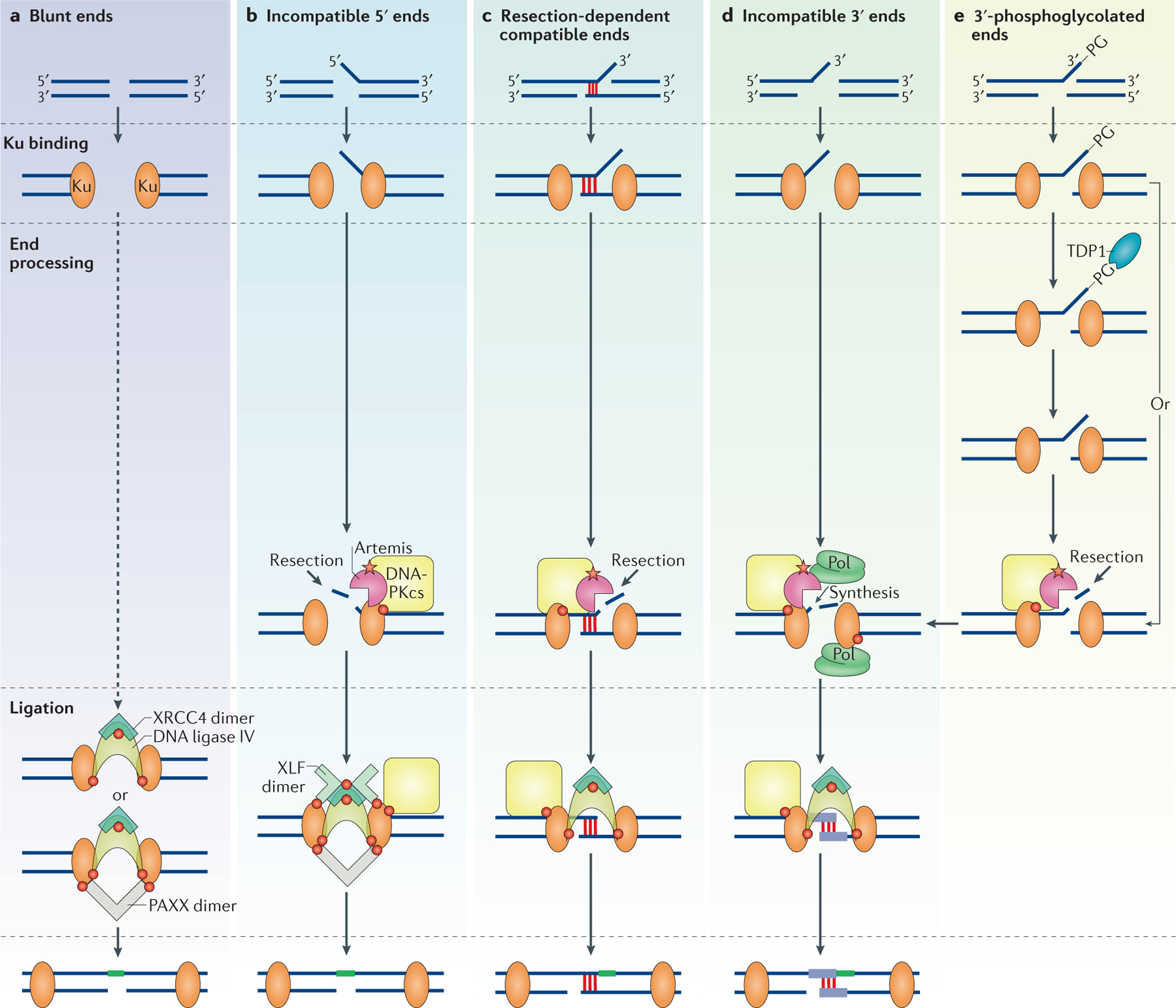
Various non-homologous end joining (NHEJ) proteins may associate at common NHEJ substrates. Red circles represent known protein–protein interactions depicted in FIG. 2. The red star represents the interaction between Artemis and DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which results in the activation of the endonuclease activity of Artemis. The diagram depicts ligation (in green) only of the top strand, but this process will inevitably proceed to the bottom strand (Supplementary information S2 (figure)). a | Blunt DNA ends are preferentially repaired without end processing and their ligation can be stimulated by paralogue of XRCC4 and XLF (PAXX). b | Incompatible 5′ overhanging ends are preferentially processed with resection of the 5′ overhang by the Artemis–DNA-PKcs complex, followed by blunt-end ligation that is stimulated by XRCC4-like factor (XLF) and PAXX. c | Resection-dependent compatible ends that have a short stretch of microhomology (~4 nucleotides of base pairing) along with a non-base paired flap only require Artemis–DNA-PKcs to cleave off the flap for ligation to occur. d | Incompatible 3′ overhanging ends are processed by iterative events of end resection and nucleotide synthesis by DNA polymerases (Pol) to generate short regions of base pairing (purple) before ligation. e | 3′-phosphoglycolated (3′-PG) ends can form on recessed or blunt ends or on a DNA overhang, and can be processed by tyrosyl DNA phosphodiesterase 1 (TDP1). Alternatively, Artemis–DNA-PKcs can bypass the 3′-PG and probably other end modifications and can endonucleolytically resect the ends that contain the modifications. Ku, Ku70–Ku80; XRCC4, X-ray repair cross-complementing protein 4.
