Figure 4 |. Double-strand break repair pathway choice.
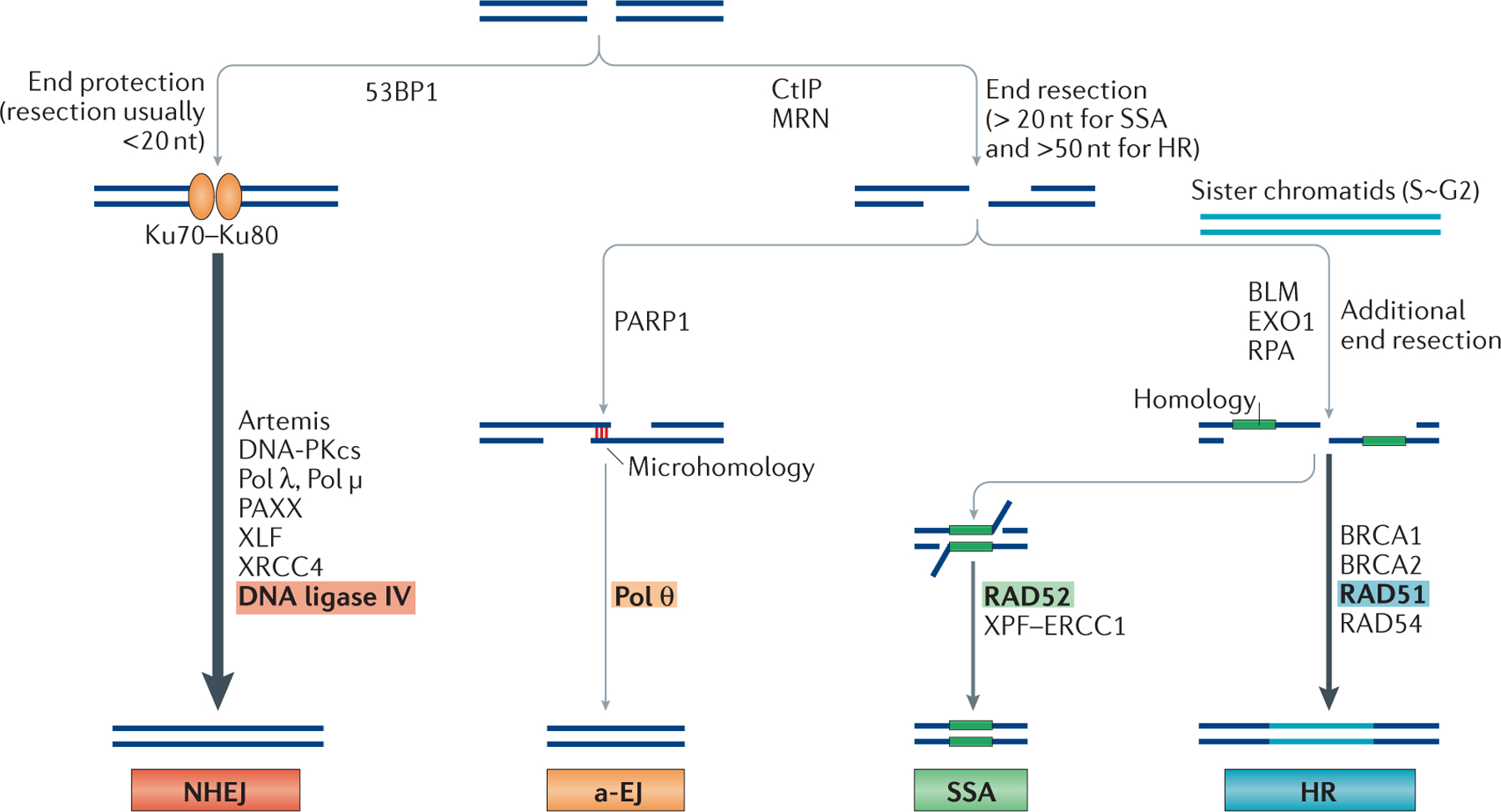
DNA double-strand breaks (DSBs) can be repaired by the classical non-homologous end joining (NHEJ) pathway, the alternative end joining (a-EJ) pathway, the single-strand annealing (SSA) pathway or by homologous recombination (HR). The major differences in pathway choice are the requirement for substantial DNA end resection. p53-binding protein 1 (53BP1) is a chromatin remodeller and a positive regulator of NHEJ. Although the complex of Artemis and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) can carry out some resection (typically <20 nucleotides), the NHEJ pathway does not require extensive end resection and the ends are mostly protected by the binding of Ku70–Ku80. By contrast, carboxy-terminal binding protein interacting protein (CtIP) and the MRN (MRE11–RAD50–NBS1 (Nijmegen breakage syndrome protein 1)) complex are involved in extensive 5′ to 3′ resection of regions of the duplex to generate stretches of single-strand DNA (ssDNA) at DNA ends for a-EJ, SSA and HR. SSA typically requires >20 bp of microhomology, whereas the requirement for a-EJ is <25 bp. Poly(ADP-ribose) polymerase 1 (PARP1) and DNA polymerase θ (Pol θ) are important for a-EJ. Bloom syndrome RecQ-like helicase (BLM) and exonuclease 1 (EXO1) provide additional resection, and replication protein A (RPA) binds to ssDNA to promote the SSA and the HR pathways. RAD52-mediated annealing of large regions of homology is key for the SSA pathway.
The xeroderma pigmentosum group F (XPF)–ERCC1 complex cuts the remaining 3′ overhangs before ligation. By contrast, RAD51-mediated strand exchange and its association with BRCA1, BRCA2 and RAD54 are essential for promoting the HR pathway. PAXX, paralogue of XRCC4 and XLF; XLF, XRCC4-like factor; XRCC4, X-ray repair cross-complementing 4.
