Abstract
The biomass demand to fuel a growing global bio-based economy is expected to tremendously increase over the next decades, and projections indicate that dedicated biomass crops will satisfy a large portion of it. The establishment of dedicated biomass crops raises huge concerns, as they can subtract land that is required for food production, undermining food security. In this context, perennial biomass crops suitable for cultivation on marginal lands (MALs) raise attraction, as they could supply biomass without competing for land with food supply. While these crops withstand marginal conditions well, their biomass yield and quality do not ensure acceptable economic returns to farmers and cost-effective biomass conversion into bio-based products, claiming genetic improvement. However, this is constrained by the lack of genetic resources for most of these crops. Here we first review the advantages of cultivating novel perennial biomass crops on MALs, highlighting management practices to enhance the environmental and economic sustainability of these agro-systems. Subsequently, we discuss the preeminent breeding targets to improve the yield and quality of the biomass obtainable from these crops, as well as the stability of biomass production under MALs conditions. These targets include crop architecture and phenology, efficiency in the use of resources, lignocellulose composition in relation to bio-based applications, and tolerance to abiotic stresses. For each target trait, we outline optimal ideotypes, discuss the available breeding resources in the context of (orphan) biomass crops, and provide meaningful examples of genetic improvement. Finally, we discuss the available tools to breed novel perennial biomass crops. These comprise conventional breeding methods (recurrent selection and hybridization), molecular techniques to dissect the genetics of complex traits, speed up selection, and perform transgenic modification (genetic mapping, QTL and GWAS analysis, marker-assisted selection, genomic selection, transformation protocols), and novel high-throughput phenotyping platforms. Furthermore, novel tools to transfer genetic knowledge from model to orphan crops (i.e., universal markers) are also conceptualized, with the belief that their development will enhance the efficiency of plant breeding in orphan biomass crops, enabling a sustainable use of MALs for biomass provision.
Keywords: bio-based economy, bio-based crops, perennial lignocellulosic crops, marginal lands, plant breeding, breeding tools
Introduction
A global bio-based economy where building-block materials, chemicals and energy are derived from biological biomass could significantly mitigate main environmental and social problems of our fossil-based society, including climate change, environmental pollution, and geopolitical tensions (McCormick and Kautto, 2013; Bennich and Belyazid, 2017). To address this challenge, more than 40 governments worldwide have explicitly set up strategies to transit toward bio-based economic systems (Dietz et al., 2018), and projections indicate that the biomass demand to sustain bio-based production chains will amount to 6.7–13.4 Bln tons year–1 in 2050, with an increase of 198–396% compared to 2011 levels (3.4 t year–1) (Piotrowski et al., 2015). Within the range of biomass sources to sustain such demand, dedicated herbaceous and woody crops have and will keep a prominent position (Piotrowski et al., 2015; OECD,, 2017). This raises concerns, as the allocation of agricultural land to biomass production conflicts with the cultivation of food crops (Dauber et al., 2012; Gelfand et al., 2013), hampering food security, destabilizing food prices, and constraining the access to food, especially to poor rural communities (Mol, 2010; Ribeiro, 2013).
To avoid conflict with food production, dedicated bio-based crops could be grown on marginal lands (MALs) (Samson and Girouard, 1998; Tilman et al., 2006, 2009; Fritsche et al., 2010; Gelfand et al., 2013; Post et al., 2013; Zhu et al., 2016; Carlsson et al., 2017; Mehmood et al., 2017), which are areas with marginal agronomic and economic potential for cultivation of food crops and currently not used by agriculture (Peterson and Galbraith, 1932; Dauber et al., 2012; Post et al., 2013; Shortall, 2013; Schmidt et al., 2015; Mehmood et al., 2017). Typical examples include abandoned lands [no longer used due to relocation of agriculture (Campbell et al., 2008)], degraded lands [no longer productive due to an intensive and unsustainable use (Daily, 1995; Campbell et al., 2008; Dauber et al., 2012)], or waste lands [with physical or environmental constraints to agriculture (Cai et al., 2010)]. MALs comprise 247–729 Mha worldwide (Hoogwijk et al., 2003; Tilman et al., 2006; Smeets et al., 2007; Campbell et al., 2008; Field et al., 2008; Haberl et al., 2010, 2011; Nijsen et al., 2012), thus retaining a great potential for growing dedicated biomass feedstocks. In fact, considering an average biomass yield of 7.9 t ha–1 from cultivation of herbaceous and woody biomass species on MALs (Nijsen et al., 2012), such acreages would be enough to supply 28–85% and 14–42% of the total biomass demand in 2050 under the 6.7 and the 13.4 Bln t scenarios, respectively.
To successfully grow bio-based crops on MALs it is critical to offset the factors that determine the marginality of these areas (Fritsche et al., 2010; Dauber et al., 2012). Most of these factors are physical and agronomic, such as adverse land morphology (e.g., steep slopes), unfavorable soil conditions (e.g., low fertility, salinity, acidity, erodibility, poor drainage, contamination by heavy metals), or hostile climate (e.g., recurrent droughts and floods, extreme temperatures) (Dauber et al., 2012; Gelfand et al., 2013; Jones et al., 2014; Meyfroidt et al., 2016; Smaliychuk et al., 2016). Others are socio-economic, as the lack of adequate transport infrastructures, undefined land ownership, or low mechanization levels and negative demographic conditions (e.g., low population density and educational level) in the regions where MALs are located (Fritsche et al., 2010; Dauber et al., 2012; Meyfroidt et al., 2016; Smaliychuk et al., 2016). Offsetting marginality through the planting of bioenergy crops requires therefore the design of cropping systems that match the site-specific conditions of each MAL at all the levels of the agricultural practice.
At the crop and agronomic level, this means adopting crops and practices that can sustain large yields of high-quality biomass (see section “Biomass Quality” for the definition of “biomass quality” in the context of biomass production on MALs) under unfavorable conditions, minimize input requirements and promote an ecological restoration of MALs (Zegada-Lizarazu et al., 2010; Dauber et al., 2012; Allwright and Taylor, 2016). Currently available crops hardly match these conditions, as they have been bred for centuries toward different targets than the production of bio-based commodities and the cultivation under MALs conditions (Trindade et al., 2010). As a result, their cultivation on MALs could even promote soil erosion and runoff of fertilizers and pesticides (Campbell et al., 2008; National Research Council,, 2008), and would likely require high external inputs to sustain optimal biomass yields (de Vries et al., 2010; Parenti et al., 2018). An array of new crops tailored to marginal environments should therefore be developed, with a central role for plant breeding. This review aims at elucidating which crops are best to be sustainably grown on MALs, which traits should be prioritized in their improvement, and which tools are effective to advance those crops and traits.
Perennial Lignocellulosic Crops: a Promising Cropping System for Biomass Production on Marginal Lands
Several studies indicate mixtures of perennial lignocellulosic biomass crops grown under low-input agriculture as suitable cropping systems for biomass production on MALs (Tilman et al., 2006, 2009; Dauber et al., 2012; Gelfand et al., 2013; Carlsson et al., 2017; Mehmood et al., 2017; Robertson et al., 2017). In this paragraph, we discuss why such systems fit well MALs conditions, and which factors are critical to ensure that their advantages are effectively delivered.
Advantages of Cultivating Mixed Perennial Lignocellulosic Crops on Marginal Lands
Mixed perennial biomass crops (MPBCs) can couple the provision of lignocellulosic biomass with the improvement or the restoration of the ecological services of MALs at all the ecosystem levels.
Regarding soil properties, MPBCs enhance soil structure and reduce erosion (Blanco-Canqui et al., 2014; Cosentino et al., 2015; Blanco-Canqui, 2016; LeDuc et al., 2017; Fernando et al., 2018), owing to a dense and prolonged soil coverage, as well as deep and branched roots (Fernando et al., 2018), which hold large amounts of water and nutrients (Blanco-Canqui, 2016; Fernando et al., 2018). This, together with the high resource use efficiency of these crops (van der Weijde et al., 2013; Lewandowski, 2016; Carlsson et al., 2017) and their low or null fertilization requirements (Tilman et al., 2006; Robertson et al., 2017; Fernando et al., 2018), determines the very low levels of nutrient leaching (especially nitrogen – N) observed in MPBCs (Glover et al., 2010; Pérez-Suárez et al., 2014; LeDuc et al., 2017; Robertson et al., 2017; Fernando et al., 2018). For example, McIsaac et al. (2010) reported that, over a 4-years comparison, unfertilized miscanthus and switchgrass leached on average 3.0 and 1.4 kg N ha–1 year–1, respectively, far lower than a conventional maize-soybean rotation (40.4 kg N ha–1 year–1). Similar conclusions have been reached in several other studies focused on different biomass perennials (e.g., poplar, willow, switchgrass, grass-legume), as thoroughly reviewed by Robertson et al. (2017). MPBCs improve also soil organic carbon (SOC) stocks (Anderson-Teixeira et al., 2009; Glover et al., 2010; Pérez-Suárez et al., 2014; LeDuc et al., 2017; Robertson et al., 2017; Fernando et al., 2018). In this regard, Chimento et al. (2016) showed that, over a 6-years trial, perennial herbaceous (giant reed, miscanthus, switchgrass) and woody (poplar, black locust, willow) species accumulated on average 45% higher SOC than continuous tilled corn in the soil portion interested by roots. This large carbon (C) sequestration results from the continuous ground coverage, the low soil disturbance, and the large rooting systems of MPBCs (Carlsson et al., 2017), and is maximized when MPBCs are established over MALs that do not store large C stocks in their soils or natural vegetation (Tilman et al., 2009; Gelfand et al., 2013). Therefore, such “low-C MALs” should be prioritized for allocation to biomass production over areas that are naturally evolving to C-rich ecosystems (e.g., forests or wetlands) (Tilman et al., 2009; Robertson et al., 2017). An incautious use of C-rich MALs for biomass production could in fact even generate a large C debt (Schulze et al., 2012; Robertson et al., 2017), that could require decades or centuries for repaying (Fargione et al., 2008; Gibbs et al., 2008; Gelfand et al., 2011).
MPBCs can also contribute to preserve water resources. Their absolute water needs per ha generally equal – or even outweigh – annual crops as maize, wheat, or sorghum (Robertson et al., 2011c, 2017; van der Weijde et al., 2013; Hamilton et al., 2015; Fernando et al., 2018), since their large biomass production and prolonged growing season implicate high evapotranspiration rates (Hamilton et al., 2015; Robertson et al., 2017). However, water use efficiencies (WUEs) of MPBCs are generally high (especially for C4 species) (Robertson et al., 2011c; van der Weijde et al., 2013; Zeri et al., 2013), which makes them best candidates for a water-effective production of biomass (Mehmood et al., 2017). This is especially true considering that several biomass perennials are drought tolerant (Mehmood et al., 2017), can be irrigated using wastewaters (Barbosa et al., 2015), and can suffice their water needs with seasonal rainfalls in temperate climates (Robertson et al., 2017). In addition, MPBCs improve water quality and water management, as their extensive roots, prolonged soil coverage, and positive effect on soil structure and porosity promote water penetration into soils (Blanco-Canqui, 2016; Fernando et al., 2018), minimizing water runoff and soil erosion (Blanco-Canqui, 2016; Acharya and Blanco-Canqui, 2018). Furthermore, the low leaching fluxes and little agrochemical needs of MPBCs minimize water pollution (Blanco-Canqui, 2016; Acharya and Blanco-Canqui, 2018). Finally, several perennial crops (e.g., willow, giant reed, miscanthus) can sequester contaminants (e.g., Cd, Pb, Zn, Cu) and pollutants from soils and water (Bandarra, 2013; Boléo et al., 2015; Costa et al., 2016), being therefore good options to remediate contaminated MALs and depurate polluted water streams (Barbosa et al., 2015; Costa et al., 2016; Acharya and Blanco-Canqui, 2018).
Biodiversity is also improved under the cultivation of MPBCs. Werling et al. (2014) showed that these cropping systems display a much wider diversity than monocultures of annual biomass crops as maize with respect to several taxonomic groups (microorganisms, arthropods, birds, and plants). These results are in line with several other studies that compared the diversity within specific biological clades in annual and perennial bioenergy crops, as Meehan et al. (2010) and Robertson et al. (2011b) (birds), Gardiner et al. (2010); Bennett et al. (2014), and Haughton et al. (2016) (insect communities), Levine et al. (2011) (methanotrophic bacteria), or Haughton et al. (2016) (plants). In addition, the specific change in land use from degraded MALs to MPBCs enhances biodiversity (Dauber et al., 2010; Meehan et al., 2010; Robertson et al., 2011a; Chauvat et al., 2014), and the enhancement is larger as the species diversity of the new cropping system is wider (Dale et al., 2010; Landis et al., 2018). The increase in biodiversity under biomass perennials boosts also fundamental ecosystem services for agriculture, as pollination (Bennett and Isaacs, 2014; Carlsson et al., 2017) or pest suppression (Werling et al., 2011; Carlsson et al., 2017), which allows for a reduced use of agrochemicals without compromising yields (Carlsson et al., 2017). Moreover, these benefits are even extended to neighboring annual croplands, where pollination, pest suppression and yields can increase up to ∼25%, thanks to the enhanced ecosystem functions of nearby MPBCs (Liere et al., 2015).
To conclude, MPBCs can also restore an economic value and an agrarian revenue to degraded lands, improving rural development. In developed countries, their cultivation contributes to diversify farm income from arable and grass lands, and offer new occupational perspectives to older or part-time farmers (Valentine et al., 2012). Alternatively, novel bioenergy production chains based on biomass from MALs can create employment opportunities in developing countries, as well as offer access to novel, clean, energy sources, which improves living and economic standards of local communities (Valentine et al., 2012).
Steps to Establish Mixed Perennial Biomass Crops on MALs
To effectively deliver the benefits promised by MPBCs, the species and the management adopted should reflect the site-specific conditions of each target MAL (Zegada-Lizarazu et al., 2010; Blanco-Canqui, 2016; Robertson et al., 2017). Therefore, the preference should go for a wide array of dedicated and locally adapted biomass perennials, each of which fitted to specific ecological niches, and globally suitable for diverse environmental scenarios (Jones et al., 2015; Robertson et al., 2017). Table 1 reports a list of possible crops. These species typically withstand well the poor MALs conditions, showing constitutive resistance to several abiotic stresses, displaying high resource use efficiencies, and requiring low inputs (Zegada-Lizarazu et al., 2010; Dauber et al., 2012; Mehmood et al., 2017). However, most of them are novel or orphan crops, which did not undergo genetic improvement so far, especially with respect to biomass-related traits (Zegada-Lizarazu et al., 2010; Dauber et al., 2012; Jones et al., 2015; Zhu et al., 2016). As a consequence, their biomass yield and quality are highly variable (Zegada-Lizarazu et al., 2010), and often considerably lower than their genetic potential (Allwright and Taylor, 2016). This is critical, as the economic viability of cultivating MPBCs on MALs largely depends on their capacity of not only surviving structural and contingent suboptimal and low-input growing conditions, but of also producing, under those scenarios, large and high-quality biomass yields (Dauber et al., 2012; Blanco-Canqui, 2016). On the one hand, high-yielding and robust varieties significantly increase farmers’ willingness of cultivating biomass perennials on MALs, by decreasing the opportunity cost of land and increasing profits (Soldatos, 2015). On the other hand, the provision of feedstocks with optimized lignocellulose composition in relation to the intended bio-based end uses (e.g., fermentation into biofuels, extraction of plant chemicals, or transformation into biomaterials as textile fibers) is critical to increase the profitability and competitiveness of the industrial use of plant biomass (Trindade et al., 2010). To enable the cultivation of MPBCs on MALs, it will be thus essential to breed varieties that couple robustness with optimal yields (Jones et al., 2015).
TABLE 1.
Promising perennial lignocellulosic crops for cultivation on MALs.
| Crop | Habit | Biomass yield (tons ha–1 y–1) | Available genetic resources to support breeding | References |
| Miscanthus (Miscanthus spp.) | Herbaceous | 15–19 | Genome sequence; genetic maps; QTLs for biomass quality, agronomic performance, plant morphology | Atienza et al., 2002, 2003a,b; Kim et al., 2012; Ma et al., 2012; Swaminathan et al., 2012; Gabrielle et al., 2014; Liu et al., 2014, 2016; Gifford et al., 2015; van der Weijde, 2016, van der Weijde et al., 2017b; Xue et al., 2016; Baute et al., 2018 |
| Switchgrass (Panicum virgatum L.) | Herbaceous | 1-22 | Genetic maps, EST databases; SNP arrays; genome samplings; BAC libraries; analyses of natural variation; QTLs for biomass, yield, plant height, flowering time, reproductive maturity | Casler et al., 2011; Gabrielle et al., 2014; Dong et al., 2015; Lowry et al., 2015; O Di Nasso et al., 2015; Serba et al., 2015; Bartley et al., 2016; Bahri et al., 2018; Baute et al., 2018; Tornqvist et al., 2018 |
| Giant reed (Arundo donax) | Herbaceous | 7-61 | Mutant and clone collections; leaf and shoot transcriptomic sequences; studies on genetic diversity (AFLP, RAPD, and microsatellites markers) | Khudamrongsawat et al., 2004; Zegada-Lizarazu et al., 2010; Pilu et al., 2014; Barrero et al., 2015; Canavan et al., 2017; Evangelistella et al., 2017; Malone et al., 2017; Valli et al., 2017 |
| Reed canarygrass (Phalaris arundinacea L.) | Herbaceous | 15 | N.A. | Pocienë et al., 2013; Lord, 2015 |
| Virginia mallow (Sida hermaphrodita) | Herbaceous | 9-20 | N.A. | Borkowska et al., 2009; Franzaring et al., 2014 |
| Cardoon (Cynara cardunculus L.) | Herbaceous | 7-15 | Genetic maps; marker arrays; QTLs for yield, biomass production, earliness | Acquadro et al., 2005; Lanteri et al., 2006, 2012; Angelini et al., 2009; Portis et al., 2009, 2012, 2014; Sonnante et al., 2011; Scaglione et al., 2012; Mauromicale et al., 2014; Martin et al., 2015; Francaviglia et al., 2016 |
| Agave (Agave spp.) | Herbaceous | 1-34 | RAPD marker array; transcriptomic sequences | Vega et al., 2001; Davis et al., 2011; Gross et al., 2013 |
| Tall wheatgrass (Thino pyrum ponticum) | Herbaceous | 3-11 | N.A. | Wang et al., 2010, 2014; Pearson et al., 2015 |
| Bamboo (Bambusa balcooa) | Herbaceous | 40-50 | N.A. | Kuehl, 2015 |
| Cup plant (Silphium perfoliatum L.) | Herbaceous | 7-13 | N.A. | Šiaudinis et al., 2015 |
| Common reed (Phragmites australis) | Herbaceous | 10-18 | N.A. | Hansson and Fredriksson, 2004; Baute et al., 2018 |
| Spanish broom (Spartium junceum L.) | Herbaceous | 18 | Studies on the genetics underneath adaptive traits for growth on steep soils | Angelini et al., 2000; Scippa et al., 2005; Di Michele et al., 2006 |
| Nettle (Urtica dioica L.) | Herbaceous | 6-15 | Expression study of SUS genes related to fiber synthesis and quality | Hartl and Vogl, 2002; Bacci et al., 2009; Backes et al., 2018 |
| Poplar (Populus spp.) | Woody tree | 7-28 | Genome sequence; genetic maps based on different types of molecular markers; QTLs for plant growth, morphology, phenology, root growth, biomass yield, cell wall quality, wood composition | Bradshaw et al., 1994; Bradshaw and Stettler, 1995; Wu, 1998; Wu et al., 2000; Cervera et al., 2001; Yin et al., 2004; Zhang et al., 2004, 2006, 2009; Tuskan et al., 2006; Gaudet et al., 2008; Rae et al., 2008; Pakull et al., 2009; Ranjan et al., 2010; Zegada-Lizarazu et al., 2018 |
| Willow (Salix spp.) | Woody tree | 5-30 | Genome sequenced; genetic maps; QTLs for growth, biomass yield, cold tolerance, drought tolerance, plant phenology, enzymatic saccharification | Hanley et al., 2002; Tsarouhas et al., 2002, 2004; Ronnberg-Wastljung et al., 2003; Rönnberg-Wästljung et al., 2005; Hanley et al., 2006; Aylott et al., 2008; Brereton et al., 2010; Zegada-Lizarazu et al., 2010; Berlin et al., 2014; Gabrielle et al., 2014; Talukder and Saha, 2017 |
| Black locust (Robinia pseudacacia) | Woody tree | 10 | N.A. | Amaducci et al., 2017 |
| Eucalyptus (Eucalyptus spp.) | Woody tree | 10-26 | Genome sequenced; transcriptomic sequences; genetic maps; QTLs for plant growth, wood quality, lignin biosynthesis, vegetative propagation | Grattapaglia and Sederoff, 1994; Grattapaglia et al., 1995, 1996; Marques et al., 1998, 2002; Thamarus et al., 2002; Kirst et al., 2004; Brondani et al., 2006; Freeman et al., 2009; Myburg et al., 2011, 2014; Zegada-Lizarazu et al., 2018 |
| Siberian elm (Ulmus pumila L.) | Woody tree | 5-19 | Chloroplast genome sequenced; breeding efforts targeting resistance to Dutch Elm Disease and wood quality | Santini et al., 2004; Geyer et al., 2007; Lamers and Khamzina, 2008; Perez et al., 2014; Zuo et al., 2017 |
| Wild tobacco (Nicotiana glauca Graham) | Woody tree | 3-9 | Expression study under drought stress; investigation of the genetic basis of metal tolerance | Curt and Fernández, 1990; Smart et al., 2001; Shingu et al., 2005 |
Target Traits and Genetic Resources to Tailor Novel Biomass Crops to MALs
The improvement of biomass yield and quality in novel species for MALs depends on the enhancement of both biomass yield and quality per se, and biomass yield and quality stability under variable abiotic conditions. This paragraph illustrates which traits are critical to achieve these equally important breeding goals, which plant ideotypes are effective to guide the improvement of such traits, and which breeding resources are currently available.
Biomass Yield
Biomass yield is a highly complex trait, overall determined by three factors: the efficiency of light interception by the crop canopy, the efficiency of light conversion into biomass, and the efficiency of biomass partitioning into target harvestable plant components (Monteith, 1972, 1977). Thus, the enhancement of biomass yield encompasses breeding for all the morphological, physiological and phenological traits that are at the basis of these three factors (Jones et al., 2015). In addition, the characteristics of vegetative tissues should deserve particular attention, as in biomass crops they not only operate light interception and conversion into biomass, but are also the main harvest product.
Efficiency of Light Interception
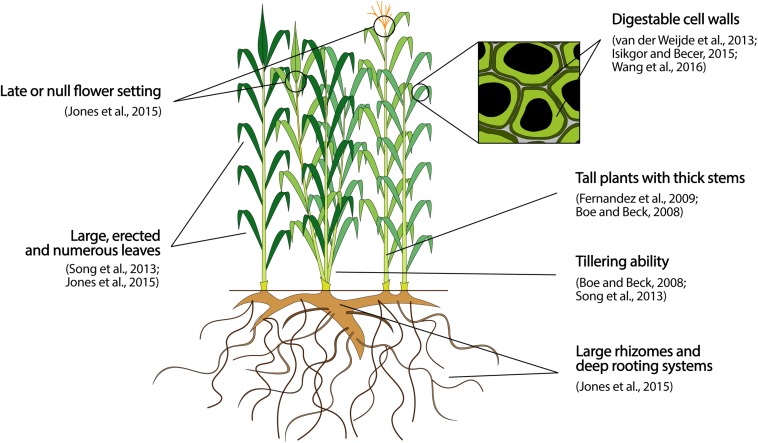
The efficiency of light interception depends on plant architecture and duration and timing of crop growth (Jones et al., 2015). Plant architecture refers to crop growth habit (i.e., plant height and branching pattern) and leaves characteristics (i.e., number, size, shape, distribution and orientation) (Reinhardt and Kuhlemeier, 2002; Hollender and Dardick, 2015; Pan et al., 2017), and affects biomass yield by determining the plant density and biomass volume achievable per land unit, as well as the degree of soil coverage and photosynthetic area of the canopy. Tall plants, high tiller/stem number and density, thick stems, and upright, large and numerous leaves are all architectural characters positively correlated with biomass yield and light penetration and interception by the canopy in herbaceous biomass crops, as shown in Figure 1 (Boe and Beck, 2008; Fernandez et al., 2009; Song et al., 2013). In woody species, vertical growth habit and production of sylleptic branches are also important to attain high plant densities and increase biomass yield per hectare (Rae et al., 2004; Marron et al., 2006; Dubouzet et al., 2013). For most of these traits, variability has been observed for crops suitable for MALs, as shown by e.g., Das et al. (2004) (switchgrass), Robson et al. (2013) (miscanthus), Cosentino et al. (2006) (giant reed), or Orlovic et al. (1998), and Rae et al. (2004) (poplar). In addition, QTLs underlying architectural traits have also been mapped in two of the most studied novel biomass species [miscanthus (Atienza et al., 2003a; Gifford et al., 2015; Ge et al., 2018), and switchgrass (Serba et al., 2015)], as well as in model biomass crops as maize (e.g., Pan et al., 2017) or poplar (Bradshaw and Stettler, 1995; Zhang et al., 2006; Rae et al., 2008). These studies highlight the high genetic complexity of plant architecture, as particularly exemplified by Pan et al. (2017), who found nearly 800 QTLs associated with ten critical maize architectural traits, including plant height, number and length of branches, and leaf number, size, and orientation. Such a complexity hampers breeding for plant architecture in novel crops for which breeding tools are largely missing. Nevertheless, major-effect architectural loci have also been identified in model energy crops as maize, sorghum, or poplar, and in the model plant Arabidopsis (see Wang and Li, 2006; Fernandez et al., 2009; and Teichmann and Muhr, 2015 for detailed reviews). These loci could potentially be targeted in candidate gene approaches, by mining eventual homologous in novel biomass crops, screening allelic diversity, and selecting or introgressing favorable alleles. Preeminent examples are the LG1 and LG2 loci of maize, that, once mutated, induce upright leaves lacking of ligules and auricles, allowing for higher planting densities (Tian et al., 2011). Alternatively, four DWARF loci (DW1-DW4) largely control internode length – and therefore plant height – in sorghum in a pure Mendelian fashion (Hilley et al., 2016; Mullet, 2017).
FIGURE 1.
Preeminent architectural, phenological, and quality target traits to breed perennial lignocellulosic biomass crops for biomass production on MALs. The traits reported represent an ideotype to guide breeding activities, and the effective magnitudes of improvement attainable with respect to each trait can vary extensively depending on the species object of a breeding program.
The duration and timing of crop growth affects biomass yield by determining both the total amount of solar radiation that can be captured during crop growth, and the duration of vegetative growth prior to switching to the reproductive cycle, which typically terminates the synthesis of vegetative tissues (Fernandez et al., 2009; Jones et al., 2015). To maximize biomass yield, an ideal crop should thus display a long growing season, fully develop the canopy by the time solar radiation reaches its yearly maximum, and postpone or even avoid the setting of reproductive structures (Figure 1) (Jones et al., 2015). Achieving this ideotype implies breeding for an early leaf development, a delayed plant senescence, and a late flowering time. As a proof-of-concept, Dohleman and Long (2009) showed that the 4-weeks earlier canopy development and the 4-weeks longer canopy duration of miscanthus compared to maize largely explain the on average 60% higher biomass production of the former in US Midwest. The genetics underlying these developmental traits are complex, as a high number of genes showing pleiotropic and epistatic effects and interacting with environment are involved (see e.g., Wu et al., 2012; Gregersen et al., 2013; Bluemel et al., 2015 for reviews). Nevertheless, some critical genes can be targeted in candidate gene or transgenic approaches. An example is the IPT gene from Agrobacterium tumefaciens that, once introgressed in several plants, boosts a rate-limiting step in the cytokinin biosynthesis and delays plant senescence by sustaining high cytokinin production along the whole growing season (Gregersen et al., 2013). Genes proven to evenly affect a critical trait across several species are undoubtedly important targets to advance novel crops without the need of de novo investigating the genetics underlying that trait in each species. However, they are generally rare for developmental traits whose genetic architecture relies on tens of small-effect loci (Buckler et al., 2009) and has been shaped by selective forces acting in a species- and environmental-specific manner during evolution, as discussed by Gifford et al. (2015) for flowering time in miscanthus. Therefore, conventional selection of superior genotypes in target MALs is nevertheless a time- and money-saving breeding approach for traits as earliness of leaf development, delayed plant senescence and late flowering time. In this regard, variation for all of them exists in several crops, as miscanthus (Farrell et al., 2006; Jensen et al., 2011; Robson et al., 2012), or switchgrass (Van Esbroeck et al., 1997; Bhandari et al., 2010). In addition, Van Esbroeck et al. (1997) and Bhandari et al. (2010) showed that the heritability of all these traits in switchgrass is moderate to high, promising success for selection. Finally, different ecotypic groups are often encountered in species that grow over wide geographical ranges [e.g., switchgrass Evans et al. (2017) or hemp Salentijn et al. (2015)], and represent an additional resource to enhance yield. For example, when short-day hemp cultivars are cultivated at northern latitudes native of long-day genotypes, flowering time is postponed and fiber yield increases (Salentijn et al., 2015).
Efficiency of Light Conversion Into Biomass
The efficiency of sunlight conversion into biomass is determined by the amount of biomass energy produced relative to the total sunlight energy intercepted by a crop over a specific period of time (Zhu et al., 2010), and depends directly on crop photosynthesis. Crops with C4 photosynthesis typically display higher conversion efficiencies than C3 species, owing to a CO2-concentrating mechanism that prevents photorespiration by sustaining high CO2 leaf concentrations for optimal Rubisco activity even at low atmospheric CO2 levels (van der Weijde et al., 2013). Such higher photosynthetic efficiency translates into higher yield potentials of C4 relatively to C3 crops (van der Weijde et al., 2013), which make the former more suitable for cultivation on MALs. This is especially true considering that C4 plants generally outperform C3 species also in terms of nitrogen and water use efficiency, given the low levels of photosynthetic proteins in leaves and low stomatal conductance (Ghannoum et al., 2011).
Promising C4 perennials for biomass production on MALs include miscanthus, switchgrass, napiergrass or Indian grass. Moreover, research is investigating how to engineer C4 photosynthesis in C3 crops (Zhu et al., 2010; Schuler et al., 2016), which could hypothetically further enhance the biomass yield of already high-yielding C3 species suitable for MALs, as giant reed or tall wheatgrass. This appears a long-term goal though (Zhu et al., 2010), as the high complexity of C4 photosynthesis and the high number of genes involved in determining all its metabolic and physiological benefits did not allow so far to achieve satisfactory results (Schuler et al., 2016).
In addition, C4 plant species evolved as a result of convergent evolution in warm, tropical, climates (Schuler et al., 2016), and C3 plants can outperform C4 species for both biomass production and resource use efficiency at temperate latitudes (Karp and Shield, 2008; Carroll and Somerville, 2009). Where this condition applies, the intra- and inter-specific variation in photosynthetic capacity that has been nevertheless observed across several C3 species (Hikosaka, 2010; Lawson et al., 2012) represents a precious resource to improve the efficiency of light conversion in promising C3 crops for MALs. For example, Wullschleger (1993) assessed the photosynthetic capacity of 109 C3 species and found carboxylation rates in a range of 6-194 μmol m–2 s–1. This variation is thought to arise mainly from different biochemical capacities of the photosynthetic machinery (e.g., antenna complex size or photosystem II photoprotection capacity), a variable degree of CO2 diffusion in leaves, different rates of N supply to the photosynthetic systems, and changes in the activity of photosynthetic enzymes (Hikosaka, 2010; Zhu et al., 2010; Lawson et al., 2012). Moreover, it appears largely genetically controlled (Zhu et al., 2010; Flood et al., 2011), even if its genetic basis remains largely understudied (Flood et al., 2011; Lawson et al., 2012; Zhu et al., 2016). Recently, QTLs for photosynthetic efficiency have been identified in model species as Arabidopsis (van Rooijen et al., 2015) or tomato (De Oliveira Silva et al., 2018). However, long time will be needed before critical genetic elements are identified underneath genomic regions associated with photosynthesis variation, and the validity of such genetic elements can be extended also to orphan crops for MALs. Nonetheless, this approach could lead to the discovery of key candidate genes to improve yield of biomass crops, and should therefore be explored in further detail.
Efficiency of Carbon Partitioning Into Vegetative Tissues
The efficiency of C partitioning refers to the amount of fixed C invested in developing vegetative tissues over the total fixed C (Zhu et al., 2010). In biomass perennials, the seasonal production of vegetative tissues is initially fueled by the C stored into roots, until the new canopy develops sufficient photosynthetic capacity to take on this role. Therefore, two main crop characteristics are critical to maximize the biomass synthesis from a C partitioning perspective (Jones et al., 2015). Firstly, a preferential allocation of fixed C to the production of vegetative tissues over other C sinks. Secondly, a rooting system that develops its full size soon after crop establishment and capable of storing large C stocks, as displayed in Figure 1.
The preferential allocation of fixed C to the production of vegetative tissues can be improved through indirect selection for architectural and phenological traits correlated with biomass yield. In this sense, plant height and late flowering time are key, as they typically correlate with each other and with biomass yield, as shown in maize (Lübberstedt et al., 1997), sorghum (Murray et al., 2008; Ritter et al., 2008), switchgrass (Bhandari et al., 2010, 2011), or miscanthus (Gifford et al., 2015). Tall plants display a relaxed C demand by sinks different than vegetative tissues thanks to a postponed switch to reproductive growth, which translates into higher relative C investments in biomass production and larger yields. Importantly, these associations appear genetically determined, as the co-localization of QTLs for plant height, flowering time, and biomass yield has also been observed, for example in sorghum (e.g., Lin et al., 1995; Murray et al., 2008; Ritter et al., 2008). Alternatively, C partitioning can also be improved through transgenic approaches, by altering the expression of sucrose synthases or sucrose transporters (Braun et al., 2013; Yadav et al., 2015). For example, Poovaiah et al. (2015) considerably increased plant height (+37%), biomass yield (+13.6%), and tiller number (+79%) in switchgrass by overexpressing a constitutive Sucrose Synthase (PvSUS1) ubiquitously present in the plant. Despite the promising results, a major drawback of these approaches is that undesired side-effects on growth or physiological traits are often encountered, which calls for a deeper understanding of the genetics underlying C allocation into a plant system perspective (Yadav et al., 2015).
Improving roots and rhizomes as determinants of C allocation in biomass crops is challenging, as studying these traits is costly, time-consuming, and technically demanding, especially in field conditions (Sartoni et al., 2015; Pierret et al., 2016). Thus, little is known on the genetics of root growth (Topp et al., 2013), especially in biomass crops. In this context, the studies on the genetic basis of rhizomatousness in sorghum using progeny from crosses of wild perennial and cultivated annual accessions (Paterson et al., 1995; Kong et al., 2015) appear particularly relevant. Following this strategy, Kong et al. (2015) mapped seven major QTLs for rhizomatousness, finding co-localizations with regions affecting tillering. The further investigation of causative genes underneath these QTLs could identify candidates to improve both rooting capacity and biomass yield components of sorghum and, possibly, other biomass perennials. In addition, novel phenotyping platforms (e.g., Nadezhdina et al., 2012; Topp et al., 2013; Sartoni et al., 2015) are expected to reduce costs and destructiveness of root phenotyping, as well as grant assessment of root growth over multi-years trials, which is needed to understand root development in a C partitioning perspective. At the moment, the most feasible strategy to breed for fast-developing and large roots is by making use of known correlations between root properties and this trait ideotype. Specifically, above-ground biomass production in MPBCs positively correlates with rooting depth (Mueller et al., 2013), which in turn affects the total C sequestration capacity of roots and rhizomes (Jones et al., 2015). Therefore, root depth appears a promising target to breed for root systems capable of storing large C stocks and sustaining abundant yields.
Biomass Quality
MPBCs produce large amounts of lignocellulosic biomass, which represents a highly attractive material for bio-based applications, as lignocellulose contains several classes of economically interesting compounds, including biopolymers and biochemicals (Trindade et al., 2010; Isikgor and Becer, 2015). The extraction of target molecules from lignocellulose is currently based on expensive and intensive post-harvest biomass treatments aimed at both loosening the structure and fractionating the components of plant cell walls, which are by far the major constituents of lignocellulose and contain the major part of attractive compounds (van der Weijde et al., 2013; Isikgor and Becer, 2015; Lauria et al., 2015; Wang et al., 2016). For this reason, the biomass quality of MPBCs entails primarily the recalcitrance of plant cell walls to deconstruction, as feedstocks with easily destructible cell walls require milder and cheaper treatments to be processed into bio-based end-products (van der Weijde et al., 2013; Isikgor and Becer, 2015; Wang et al., 2016). Moreover, the relative content of molecules of interest within cell walls in relation to the end-use of the biomass is also a preeminent quality target in order to develop crop varieties tailored to specific bio-based production chains (Trindade et al., 2010). In this regard, Table 2 reports a list of cell wall ideotypes that can fit the needs of different possible end-uses of the lignocellulosic biomass obtainable from MPBCs grown on MALs.
TABLE 2.
Possible end-uses of the lignocellulosic biomass obtainable from MPBCs grown on MALs and relative cell wall ideotypes to optimize biomass quality toward those applications.
| Biomass application | Optimal cell wall ideotype to increase the biomass quality toward target applications | References |
| Combustion for heat or energy generation | High lignin/low cellulose and hemicelluloses relative contents to increase the caloric value of the biomass. Low amounts of mineral components to avoid ash formation and corrosion of combustion chambers upon burning. | van der Weijde et al., 2017c |
| Production of bioethanol | Lignin: low relative content, low degree of cross-linking with cellulose and hemicelluloses. Cellulose: high relative content, low degree of polymerization, low crystallinity index. Hemicelluloses: high relative content, low degree of branching with cellulose and lignin (largely achievable through a low rate of xylan substitutions). | van der Weijde et al., 2013; Torres et al., 2015b; da Costa et al., 2019 |
| Production of textile fibers | Lignin: low relative content. Cellulose: high relative content, high crystallinity index, small microfibril angles. Hemicelluloses: low rate of xylan substitutions to decrease cross-linking with cellulose, lignin and pectins. Others: low rates of pectin methylation, low cross-linking with other structural cell wall components. | Ranalli, 2004; Salentijn et al., 2015 |
| Biorefining into polymers/chemicals | Lignin: high relative content for production of aromatic molecules of nutraceutical, cosmetic, pharmaceutical, and chemical interest, otherwise low relative content; tailor relative contents of S, G and H monolignols to the desired properties of end-products. Cellulose: high relative content for production of glucose and related functionalized derivatives; low crystallinity index. Hemicelluloses: high relative content for production of C5 (xylose, arabinose) and C6 (mannose, galactose, rhamnose) sugars and related functionalized derivatives to be used as polymer backbones or pendant groups in chemical industries; altered relative abundance of the different hemicelluloses polysaccharides based on the wishes of end-users. Others: optimal design of the cross-links between structural cell wall components and of the deposition of cell wall molecules within cell walls to obtain specific properties in target molecules and/or facilitate extraction and processing. | Isikgor and Becer, 2015; da Costa et al., 2019 |
The general structure of cell walls is conserved across plants, consisting of cellulose fibers in a matrix of non-cellulosic polysaccharides (mainly hemicelluloses), lignin, structural proteins and mineral elements (Cosgrove, 2005; Vogel, 2008). However, the relative abundance, composition, and structure of cell wall components vary extensively across species, tissues and developmental stages (Vogel, 2008; Sarkar et al., 2009; Loque et al., 2015). Likewise, the occurrence and functionality of major cell wall synthetic genes is also conserved across plants (e.g., CESAs and CSLs as main players of cellulose and hemicellulose biosynthesis, respectively) (Vogel, 2008; Xu et al., 2009; Zhang et al., 2018), even if inter-specific differences exist also at the genetic level, which affect cell wall composition (e.g., the presence of CSL-Fs and CSL-Hs only in certain plant clades, which synthesize mixed-linkage glucans) (Vogel, 2008; Doblin et al., 2009). Taken together, these observations point out a large margin for the improvement of cell wall composition toward low-recalcitrant and purpose-made cell wall ideotypes. However, the extreme complexity of cell wall biosynthesis and regulation [∼4000 genes are thought to be involved in Arabidopsis (Wang et al., 2012)] hampers breeding efforts, especially in novel crops lacking genetic tools. In this context, dissecting the trait “cell wall quality” into its main determinants (content, composition, and structure of lignin and cell wall polysaccharides) can help to better define the goals targetable by breeding, and the available biochemical and genetic knowledge to achieve them.
Lignin
Lignin is the cell wall component that mostly limits lignocellulose deconstruction (Li et al., 2016). On the one hand, it cross-links with hemicelluloses forming a physical barrier that hides polysaccharides to degrading enzymes (Zhao et al., 2012; Li et al., 2016). On the other hand, it irreversibly absorbs hydrolytic enzymes, inhibiting their activity (Zhao et al., 2012; Li et al., 2016). To decrease biomass recalcitrance, an immediate strategy is to decrease the lignin content in cell walls (van der Weijde et al., 2013). However, as lignin provides mechanical support, stress response, and pathogen resistance to plants, decreased lignin contents can hamper plant stability and growth, and ultimately reduce biomass yield (Wang et al., 2016). Moreover, lignin itself represents an economically attractive compound, as it is a source of aromatic molecules that can find applications in the production of phenolics, carbon fibers, dispersants, and bio-plastics (Isikgor and Becer, 2015). Therefore, strategies aimed at modifying the lignin properties that affect biomass digestibility without decreasing the total lignin content – such as altering ratios of lignin subunits, relocating lignin deposition, and modifying lignin backbone and linkages with polysaccharides – also represent valid breeding approaches to improve biomass quality (Verma and Dwivedi, 2014; Li et al., 2016; da Costa et al., 2019).
The lignin biosynthetic pathway is well characterized (Boerjan et al., 2003; Bonawitz and Chapple, 2010) and also highly conserved across vascular plants (Boerjan et al., 2003; Loque et al., 2015). These characteristics make candidate gene approaches particularly suitable to modify lignin properties, by identifying critical target genes within the lignin pathway, testing their effectiveness in model crops, and transferring successful approaches to other less-studied species, as orphan biomass perennials. For example, the relevance of down-regulating the Caffeoyl O-Methyltransferases (COMTs) to decrease lignin content and improve biomass digestibility for biofuel production has been firstly shown in model forage crops as maize, tall fescue, and alfalfa (Hisano et al., 2011). These results have pushed the successful reproduction of this approach in switchgrass (Fu et al., 2011), while gene cloning and in silico protein alignments led to the identification of COMT genes in miscanthus (Dwiyanti et al., 2014) and eucalyptus (Carocha et al., 2015), which can become future targets of genetic modification to improve biomass quality. According to this approach, the improvement of lignin properties in novel lignocellulosic perennials for MALs can benefit from the long list of successful modifications of lignin-related genes in model plants and staple biomass crops (see Liu et al., 2018 for a recent overview). Overall, these studies also highlight four important principles for lignin modification. First, lignin content is decreased more effectively when genes acting early in lignin biosynthesis are targeted (Chen and Dixon, 2007). Second, down-regulation of genes is more effective than knock-outs to minimize side-effects on plant growth (Xie and Peng, 2011). Third, pathway cross-talks and gene redundancy need to be carefully considered to exclude mechanisms that can limit the gains from targeted approaches (Torres et al., 2015b). Fourth, to reproduce successful transgenic approaches in novel biomass crops, the availability of transformation and regeneration protocols is critical (Clifton-Brown et al., 2018).
Besides genetic modification, large natural variation in lignin content and composition exists within biomass perennials promising for MALs, as miscanthus (Zhao et al., 2014), switchgrass (Yan et al., 2010), or willow and poplar (Kenney et al., 1990). Moreover, studies investigating the sources of such variability showed that it is typically highly heritable (Torres et al., 2015a; van der Weijde et al., 2017b), and therefore constitutes an important breeding resource. Alternatively, intra-specific crosses between accessions showing contrasting lignin profiles have been also successfully used to map lignin-related QTLs across a range of species (Thumma et al., 2010; Torres et al., 2015a; van der Weijde et al., 2017b), which open prospects for marker-assisted selection (MAS) of genotypes showing superior lignin profiles and high biomass degradability.
Cell Wall Polysaccharides
Cellulose and hemicelluloses represent attractive polymers for bio-based applications, as they constitute the bulk of energy contained into lignocellulosic biomass and can be used as platforms to produce several classes of valuable biochemicals (van der Weijde et al., 2013; Isikgor and Becer, 2015). Therefore, increasing the cellulose and hemicellulose content in cell walls and modifying their molecular properties that promote recalcitrance are also effective targets to improve biomass quality (Torres et al., 2015b). Cellulose recalcitrance depends on the degree of cellulose crystallinity and polymerization (Wang et al., 2016), and reducing these two parameters is critical to improve lignocellulose degradability (van der Weijde et al., 2013; Torres et al., 2015b; Allwright and Taylor, 2016). Conversely, hemicelluloses affect recalcitrance through their total content in cell walls and their degree of branching (Torres et al., 2015b; Wang et al., 2016). Specifically, as hemicelluloses cross-link cellulose and lignin, high hemicellulose content reduces cellulose crystallinity (Wang et al., 2016). At the same time, low xylan branching ensures an easy separation of hemicelluloses, cellulose and lignin during saccharification (Torres et al., 2015b).
The molecular alteration of cell wall polysaccharides is challenged by our limited knowledge on cellulose and hemicellulose biosynthesis and regulation. Research in this field is still at the level of functional genetics in model species as Arabidopsis, while attempts of candidate gene studies in biomass crops remain few, mainly restricted to poplar and – to a less extent – maize (Wang et al., 2016). Several studies assessed the effect of modifying cellulose synthases (CESAs) on cellulose properties. For example, Joshi et al. (2011) decreased cellulose crystallinity in poplar wood by overexpressing a constitutive CESA (PtdCESA8). Alternatively, Harris et al. (2009) mutated the AtCESA3, which led to a 34% reduction of cellulose crystallinity and a 151% increase of biomass degradability. Besides CESAs, numerous other genes affect the molecular properties of cellulose and hemicelluloses. Glass et al. (2015) reduced cellulose crystallinity and increased biomass yield in Arabidopsis by down-regulating a class C endoglucanase (AtGH9C2). Furthermore, Mortimer et al. (2010) mutated the two glycosyltransferases-coding loci GUX1 and GUX2 in Arabidopsis, achieving substitutions-free hemicelluloses and easily extractable xylans. Overall, these studies highlight promising targets to improve cell wall polysaccharides, for which homologs could be searched in biomass crops and considered for genetic modification or screening of allelic diversity for conventional breeding programs (Torres et al., 2015b). However, in order to better predict/avoid the (negative) pleiotropic side-effects that are often encountered in these studies, it is critical to develop a better, systemic, understanding of the genetic complexity of cellulose and hemicellulose biosynthesis and regulation, and their interplay with other plant metabolic pathways. For example, along with the decreased cellulose crystallinity, the poplar transgenic lines produced by Joshi et al. (2011) displayed also a decreased cellulose content, increased lignin, and stunt growth, which are all unwanted traits for elite lines of perennial biomass crops.
As for lignin, variability in the composition and structure of cell wall polysaccharides across species constitutes an important breeding resource. In this regard, Harris and Debolt (2008) reported large variation in cellulose crystallinity across a set of 35 plant species, while Vandenbrink et al. (2012) and Porth et al. (2013) found considerable intra-specific variation in sorghum and poplar association panels, respectively. Alternatively, Torres et al. (2013) found variable hemicellulose composition and substitution patterns in a maize doubled-haploid population, which correlated with differences in biomass digestibility. To conclude, when molecular markers are available or can be easily developed, variability can also be used for association mapping, leading to the identification of genomic regions associated with high biomass quality, as performed by Torres et al. (2015a) in maize or van der Weijde et al. (2017b) in miscanthus.
Biomass Yield and Quality Stability Under Abiotic Stresses
Abiotic stresses as water surplus or deficit, extreme temperatures, and soil salinity are common constraints to agriculture on MALs, and are expected to intensify as a result of climate change (Jones et al., 2015; Quinn et al., 2015). These stresses discourage the cultivation of MALs, as they can hamper crop growth, reduce biomass yield and quality, and ultimately hinder a stable biomass supply, which would volatilize prices of raw biomass and bio-based commodities. Developing genotypes that show stability of biomass yield and quality under adverse abiotic conditions is thus pivotal to successfully allocate worldwide MALs to biomass production. Specifically, since abiotic stresses often occur in combination (e.g., heat and drought) or succession (e.g., waterlogging followed by drought) (Mickelbart et al., 2015), resistant varieties should possibly combine different sources of resistance to withstand multiple stresses at once.
Suboptimal Water Availability
Water shortage inhibits cellular expansion, hydration and photosynthesis, with negative impacts on plant germination, establishment, growth, nutrient assimilation and transport (Quinn et al., 2015), and ultimately biomass yield (Emerson et al., 2014). Flooding determines instead stomata closure and uptake of toxic compounds released by anaerobic microorganisms in anoxic soils, which inhibit nutrient transport and photosynthesis, damaging plant growth and yield (Quinn et al., 2015). Drought and waterlogging are major causes of yearly yield losses worldwide and are acquiring more-than-ever importance as a consequence of extreme atmospheric events in a changing climate (Jones et al., 2015). Moreover, common MALs characteristics as steep slopes, high erodibility, or poor drainage amplify the occurrence and effects of these stresses (Quinn et al., 2015), calling for crops able to maintain normal metabolism, growth, and yield under drought and flooding.
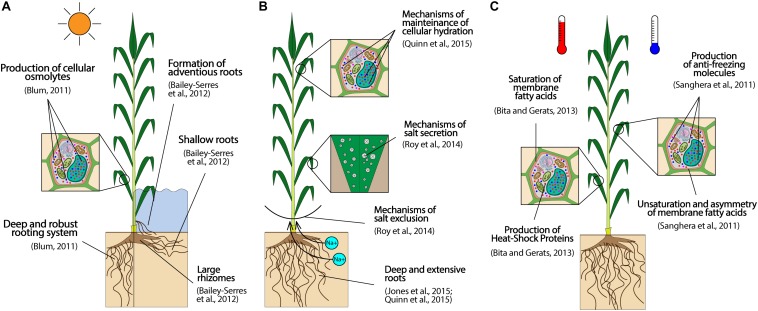
Several plant traits can be targeted to develop drought- and flood- tolerant crops, as visually summarized in Figure 2A. For drought, deep and robust roots, able to penetrate harsh soils, are important characters to reach deep water in dry areas (Blum, 2011). Moreover, the ability of accumulating cellular osmolites to avoid dehydration (osmotic adjustment) is also important, particularly at the seedling stage when roots are still underdeveloped (Blum, 2011), and can be improved by selecting against leaf rolling and in favor of green canopy under drought, which are two easily scorable traits (Amelework et al., 2015). For flood tolerance, a shallow root system, with a thick root epidermis, well-developed aerenchymatous tissues, and adventitious roots are critical traits as they facilitate aeration (Bailey-Serres et al., 2012). Moreover, large rhizomes are also favorable to provide starch to sustain optimal growth under prolonged flooding (Bailey-Serres et al., 2012).
FIGURE 2.
Preeminent target traits to equip perennial lignocellulosic biomass crops with effective resistance to common abiotic stresses of MALs: drought (A, left), flooding (A, right), salinity (B), and extremely warm (C, left) or cold (C, right) temperatures. The traits reported represent an ideotype to guide breeding activities, and the effective magnitudes of improvement attainable with respect to each trait can vary extensively depending on the species object of a breeding program.
Natural variation in tolerance to suboptimal water conditions is an important breeding resource, especially considering the high polygenicity of this trait (Mickelbart et al., 2015), and the difficulty of phenotyping roots in field conditions (see section “Efficiency of carbon partitioning into vegetative tissues”). Different WUEs between species are a first important variable trait, given the direct correlation between WUE and drought tolerance (Allwright and Taylor, 2016). In this regard, as previously discussed (see section “Efficiency of light conversion into biomass”), C4 crops display overall higher WUEs than C3 species, and are thus best options on dry MALs. In addition, intra-specific variation in the capacity of undergoing drought or floods without substantial yield penalties has been observed across several biomass perennials. For example, Barney et al. (2009) showed that upland and lowland switchgrass ecotypes display significantly different levels of drought tolerance. This observation is relevant, as it could enable to simultaneously improve drought tolerance and other traits displaying analogous latitudinal variation, such as flowering time, biomass yield, and cold resistance (Clifton-Brown et al., 2018). In miscanthus, van der Weijde et al. (2017a) have found large variation in the capacity of 50 accessions to produce biomass under drought, as their drought-constrained yields amounted to 30–110% of the yields under control conditions. The study assessed also biomass degradability under drought, demonstrating that it increased across all the 50 accessions, irrespectively of the level of drought tolerance, probably through an increase of the hemicellulose/cellulose ratio in cell walls (van der Weijde et al., 2017a). This is a remarkable finding, as it potentially allows to develop genotypes that are not only drought-tolerant, but also capable of turning the drought burden into an advantage for biomass quality. Finally, in black locust, Zhang et al. (2010) observed that tetraploid accessions withstand better drought than diploid genotypes, an information that can be useful at the moment of choosing superior lines to be used as parents in breeding programs (even if polyploidy can complicate breeding).
A better understanding of the genetics underlying drought and flood tolerance paves also the way to improve these traits through candidate gene approaches. Transcription factors (TFs) related to hormone metabolism are consistently accumulated under both drought and flooding across a range of plants (Yin et al., 2014; Mickelbart et al., 2015), and appear thus relevant to mine candidate genes. For example, Yang et al. (2015) and Zhao et al. (2016) enhanced drought, cold, and salt tolerance in Arabidopsis by ectopically overexpressing two NAC TFs from Miscanthus lutarioriparius (MlNAC5, MlNAC9) involved in the ABA-dependent signaling pathway. These results highlight the relevance of these genes to equip Miscanthus (and possibly other perennial grasses) with tolerance to multiple abiotic stresses, and their validity should be now investigated in target biomass crops. Alternatively, ERF-VIIs are TFs upregulated upon flooding across several plants (Mickelbart et al., 2015). They include the rice SUB1A gene, which provides flood tolerance to rice by quitting growth upon flooding, limiting resource dispersal (Xu et al., 2006). Manipulating the expression of ERF-VII TFs could thus be key to provide flood tolerance (Mickelbart et al., 2015), and deserves deeper investigation, especially in biomass crops. In fact, even if proper flood tolerance has not yet been improved through this approach (Peña-Castro et al., 2011), the expression of OsSUB1A in Arabidopsis not only led to flowering inhibition (mimicking the quiescence survival strategy observed in rice) (Peña-Castro et al., 2011), but also improved the saccharification efficiency of the biomass (Nunez-Lopez et al., 2015). Therefore, possibilities to improve both flood tolerance and biomass quality by manipulating ERF-VIIs could be furtherly explored. More in general, the interplay between cell wall quality and tolerance to abiotic stresses could be key to breed biomass crops, as cell wall composition and expression of cell wall genes are deeply changed under abiotic stresses, including drought and flooding (Le Gall et al., 2015; Houston et al., 2016). Unraveling genes acting upstream of both cell wall changes and induction of mechanisms of abiotic stress tolerance could thus represent a promising strategy to breed for both these traits in parallel.
Extreme Temperatures
Freezing (<−1°C), chilling (0–18°C), or high (>35–45°C, depending on species sensitivity) temperatures can physiologically and physically damage plants, penalizing biomass yield (Quinn et al., 2015). Cold temperatures reduce germination rates, growth, and tillering (Yadav, 2010), which delay or hamper biomass production (Jones et al., 2015). Moreover, cell wall composition is also affected by frost, probably with negative effects on biomass recalcitrance, as lignin content is typically increased (Le Gall et al., 2015). Heat stress shortens the growing season by accelerating feedstock maturation and inhibits photosynthesis, causing sugar catabolism (Quinn et al., 2015). Thus, both biomass yield (Bita and Gerats, 2013) and quality (Ananda et al., 2011) are penalized.
Tolerance to extreme temperatures starts by cultivating crops adapted to the temperature ranges of a target area. In fact, while temperate species – like many C3 crops (Jones et al., 2015) – are capable of cold acclimation to withstand chilling temperatures without impacts on crop performance, tropical and subtropical plants typically lack of such mechanisms, showing sensitivity even to chilling conditions (Sanghera et al., 2011). Conversely, crops from tropical and subtropical regions – such as the majority of C4 species (Schuler et al., 2016) – withstand better heat stress (van der Weijde et al., 2013), and could thus be preferentially planted in warm environments, where could eventually display combined tolerance to heat and drought. Once crops are correctly allocated, plant breeding can enhance freezing and heat tolerance to species already adapted to cool or warm environments, respectively. To this aim, high unsaturation and asymmetry in membrane lipids (which lower the membrane freezing point), and production of protective molecules (antioxidants, chaperones, deposition of apoplastic sugar) are important traits to prevent cellular damages from freezing (Figure 2C, right) (Sanghera et al., 2011). Conversely, heat tolerance is enhanced by production of heat-shock proteins (HSPs), secondary metabolites to protect against oxidative damage, and membrane saturated fatty acids to increase membranes melting point (Figure 2C, left) (Bita and Gerats, 2013).
Because of the complex molecular and genetic nature of the traits just mentioned, selection of tolerant accessions within pools showing variability for cold and heat resistance represents an effective breeding strategy (Bita and Gerats, 2013; Jones et al., 2015). In this respect, measuring electrolyte leakage following freezing is a rapid screening method for frost tolerance (Jones et al., 2015). Vice versa, assessing morphological traits that reduce heat load [e.g., pubescent, vertically oriented, or light-colored leaves (Quinn et al., 2015)], as well as physiological characters as maintenance of photosynthesis, chlorophyll content, and stomatal conductance under heat stress (Bita and Gerats, 2013) allows quick screening of heat tolerance. Furthermore, variability for both cold and heat tolerance has been observed for several biomass perennials (Quinn et al., 2015). For example, the level of frost tolerance is variable across diverse miscanthus genotypes (Clifton-Brown and Lewandowski, 2000; Farrell et al., 2006), and cold-tolerance could be coupled with breeding for earlier germination, which is also variable in miscanthus (Clifton-Brown and Jones, 1997) and is a target trait to increase biomass yield (see section “Efficiency of light interception”).
Besides selection within intra-specific variation, sources of both heat and cold tolerance can be also introgressed through crosses with wild relatives evolved under stressed environments, or by transgenic modification. The first approach has been successfully applied in wheat (Mohammed et al., 2014), and could be used in biomass crops in which different species show different levels of tolerance to extreme temperatures, as poplar [Populus euphratica is tolerant to both heat and drought, and candidate genes and mechanisms underneath tolerance have been identified (Ferreira et al., 2006; Tang et al., 2013)] or sugarcane [the wild species Saccharum spontaneum shows higher cold tolerance than the most cold tolerant commercial varieties of sugarcane (Hale et al., 2014)]. The second approach has instead been successfully applied in eucalyptus, where the transgenic introgression of CBF genes [a class of TFs that promote cold acclimation across a range of plants (Mickelbart et al., 2015)] has greatly improved cold tolerance (Hinchee et al., 2011). As several genes involved in cold and heat tolerance are known across plants (see Sanghera et al., 2011 for a review), this approach could be reproduced also with other species and other targets.
Salinity
Soil salinity hampers plant growth as it reduces osmotic potential – which challenges water uptake and solute movement – and causes ion cytotoxicity as salt ions compete with potassium to occupy enzymatic active sites (Mickelbart et al., 2015; Quinn et al., 2015). In biomass crops, salinity consistently decreases biomass yield (Quinn et al., 2015) and remodels cell wall composition, which is thought to negatively affect biomass quality, as lignin content is typically increased (Le Gall et al., 2015).
Not all biomass crops are salt sensitive, and the allocation of tolerant species to salty MALs is a preeminent strategy to avoid salt-induced penalties to biomass production (Jones et al., 2015). In this regard, some herbaceous biomass perennials as giant reed or Pennisetum purpureum display striking levels of salt tolerance, comparable with halophytes (Quinn et al., 2015). Furthermore, biomass trees as Eucalyptus camaldulensis, several poplar and willow hybrids, and tetraploid Robinia pseudacacia accessions also show high tolerance to salinity (Quinn et al., 2015). Globally, all these species represents good options for salty MALs.
Crops can also be equipped with mechanisms of salt tolerance through plant breeding. Since salty and dry soils are functionally similar, plant traits benefiting drought tolerance are also effective to improve salt tolerance (Quinn et al., 2015). Specifically, deep and large root systems and the capacity of maintaining cellular hydration appear particularly critical characters, as shown in Figure 2B. These traits allow plants to search for water and nutrients in deep soil layers less affected by salinity, and to keep water-to-salt ratios in cells at acceptable levels (Jones et al., 2015; Quinn et al., 2015). In addition, mechanisms of salt exclusion from shoots, and secretion of excess salt uptake are other effective characters to provide salt tolerance (Figure 2B) (Roy et al., 2014), and can be effectively phenotyped by screening ion levels in plant tissues (i.e., total Na+ and Cl– contents and K+/Na+ ratio) (Chen, 2018). For example, Chen (2018) has successfully applied this method to screen levels of salt tolerance across 70 miscanthus genotypes, revealing a relatively high degree of genetic diversity in the ability of withstanding salinity among the accessions. The study has specifically showed the presence of genotypes capable of excluding salt ions from shoots, preventing leaf senescence, and sustaining biomass production on salty soils (Chen, 2018). These genotypes could be used as parents in future miscanthus breeding programs (Chen, 2018). Besides miscanthus, genetic variability for biomass production under salinity has been observed also in other biomass perennials, as switchgrass (Liu et al., 2014), black locust (Wang et al., 2013), or poplar (Sixto et al., 2005), and could be used for breeding. However, the assessment of the capacity of maintaining biomass quality under salinity – in addition to biomass yield – is often skipped by studies investigating salt tolerance. This is a major shortcoming that can lead to negative selection biases (i.e., selection of salt tolerant genotypes that eventually turn out to be highly recalcitrant to biomass processing), as studies in miscanthus have shown that best-performing accessions for salt tolerance do not coincide with genotypes showing highest biomass quality under salt stress (Chen, 2018).
Tools and Strategies to Advance Promising Lignocellulosic Perennial Crops for MALs
Available Breeding Tools
Until now, only a limited set of lignocellulosic biomass crops promising for MALs have undergone breeding (Allwright and Taylor, 2016), while the majority lies in a state close to (selected) wild germplasm (see Table 1). This small set of species includes miscanthus, switchgrass, willow, poplar, and eucalyptus, whose improvement history dates back to the second half of the twentieth century (Grattapaglia and Kirst, 2008; Allwright and Taylor, 2016; Clifton-Brown et al., 2018). All these species are outcrossing, and until early 2000s their genetic improvement has entirely relied on phenotypic selection within breeding populations created through recurrent inter-mating of wild and advanced germplasm, or intra- and inter-specific hybridization (Grattapaglia and Kirst, 2008; Clifton-Brown et al., 2018). These methods allow to combine favorable traits from distant genetic pools into elite lines and to exploit heterosis (given that the genetic structure of target populations and heterotic compatibility are analyzed prior to design breeding schemes) (Acquaah, 2012). However, the release of commercial varieties takes long time (i.e., 11–26 years), which significantly delays the adoption into real agricultural contexts of the improvements achieved in breeding programs (Clifton-Brown et al., 2018). This is due to the long period (sometimes several years) that perennial crops often require to phenotypically express traits of interest, the long breeding cycles (especially in biomass trees), and the prolonged time to commercially upscale elite lines of highly heterozygous outcrossing species (Clifton-Brown et al., 2018).
Over the last decades, research investments have permitted to develop advanced tools to assist the improvement of these crops at all the breeding levels (Clifton-Brown et al., 2018). The first tools created were genetic maps which, together with the use of mapping populations segregating for a target character that is phenotypically divergent between the founder individuals, have enabled to map quantitative trait loci (QTLs) for growth, quality, and agronomic traits (see Table 1 for relevant references). Initially, QTL detection has exclusively relied on bi-parental crosses, but more recently multi-parental approaches to mine polymorphisms linked to loci of interest – which allow to enhance the allelic diversity and variety of genetic backgrounds included in a study – have begun to be applied also in biomass crops (Mandrou et al., 2014). The availability of markers and QTLs has in turn enabled marker-assisted selection (MAS) (Clifton-Brown et al., 2018). MAS encompasses genotyping breeding material for the alleles harbored at marker loci associated with QTLs of interest, and selecting accessions carrying positive alleles at those marker loci. Therefore, selection can take place already at early developmental stages, even before a trait is phenotypically expressed, which can significantly accelerate breeding gains. For example, Thavamanikumar et al. (2018) applied MAS to improve wood density, pulp yield, and total plant growth in eucalyptus, showing that it can reduce by 50% the duration of breeding cycles (from 10–15 to 5–7 years), while breeding gains can be achieved at a 2–3 fold higher rate than by conventional selection.
The drop of genotyping costs brought by genotyping-by-sequencing (GBS) technologies, coupled with the release of the genome sequences of all the species referred in here [Tuskan et al. (2006) (poplar), Myburg et al. (2014) (eucalyptus), Dai et al. (2014) (willow)1, (miscanthus, switchgrass)] has permitted the development of dense arrays of single nucleotide polymorphisms (SNPs) covering the whole genome, which have in turn paved the way to genome-wide association studies (GWAS) and genomic selection (GS) schemes (Allwright and Taylor, 2016; Clifton-Brown et al., 2018). GWAS is a powerful tool to detect marker-traits associations using genotypic collections inclusive of long recombination histories, which promises to achieve deep mapping resolutions, and save the time needed to set up experimental populations for QTL mapping (Bernardo, 2016). Because of their high genetic diversity, undomesticated status, and generally fast linkage disequilibrium (LD) decay, lignocellulosic crops – especially biomass trees – appear ideal for GWAS (Du et al., 2018), and several studies have thus used this approach to reveal loci underlying biomass-related traits. For example, GWAS has been used in poplar to detect several marker-trait associations for quality characters as lignin content and composition (Porth et al., 2013), as well as for phenology traits as canopy duration or flowering date (McKown et al., 2014). Despite GWAS promises, Fahrenkrog et al. (2017) have pointed out how rare allele variants, whose detection can be quite often missed by GWAS analyses (Bernardo, 2016), can be particularly relevant to explain genetic variation for bioenergy traits as cell wall composition. Therefore, good experimental designs (e.g., adequate sample size and geographical sampling of accessions to give a balanced representation of the variability for a trait of interest in the panel used; Brachi et al., 2011) are pivotal to successfully perform a GWAS (Du et al., 2018). As for QTL mapping, GWAS results can be directly used for MAS (Allwright and Taylor, 2016). However, genome-wide marker allelic effects from GWAS analyses can also be used to calculate breeding values for every individual plant in a breeding population, which is the concept standing behind genomic selection (GS) (Heffner et al., 2009). GS is particularly suited for crops showing large phenotypic and genetic variability, as miscanthus (Allwright and Taylor, 2016), where the feasibility of applying this strategy has started to be explored (Davey et al., 2017). To conclude, marker arrays are also useful to screen the genetic diversity of novel germplasm collections, which is a common need of pre-breeding research in orphan crops (Clifton-Brown et al., 2018). Diversity screenings can be informative to establish the geographical origin and the relatedness with other plant material, which are important information to take breeding decisions (Narasimhamoorthy et al., 2008; Lu et al., 2013).
Research on miscanthus, switchgrass, poplar, willow and eucalyptus has also aimed at developing transformation protocols to insert genes underlying traits hardly found in extant accessions. Clifton-Brown et al. (2018) and Kendurkar and Rangaswamy (2018) have recently reviewed the progress achieved in this field across these five crops, highlighting that stable protocols are available for switchgrass, miscanthus, and poplar, while willow and eucalyptus can display recalcitrance to transformation. In addition, several studies demonstrated the efficacy of genetic modification to improve traits for which critical candidate genes are known, as discussed in section “Target traits and genetic resources to tailor novel biomass crops to MALs.” Overall, the public acceptance of genetic modification for biomass crops grown for bio-based applications could be higher than that for food crops (van der Weijde et al., 2013). However, when transgenic lines of outcrossing species were effectively cultivated, measures of gene confinement should be designed, as (trans)gene flow to relative wild species could be an issue (Clifton-Brown et al., 2018).
To conclude, fast and cost-effective phenotyping is also an asset to improve understudied crops, for which screening large germplasm collections is fundamental to evaluate variability for breeding programs (Clifton-Brown et al., 2018). Recent advances in high-throughput phenotyping open promising prospects in this regard. Fernandez et al. (2017) developed a robotic workstation that can be used to phenotype yield-related traits in tall herbaceous biomass crops as sorghum. The system has been successfully used to phenotype stem diameter and plant height in a GWAS sorghum panel, and the data collected allowed the detection of known QTLs for these traits, demonstrating the efficacy of this platform (Fernandez et al., 2017). Near-infrared spectroscopy (NIRS) technologies offer instead a viable option for high-throughput phenotyping of cell wall compositional traits, and robust protocols for their application have been recently developed and successfully used to phenotype a mapping population of miscanthus (van der Weijde et al., 2017b). Finally, thermal aerial imaging constitutes a high-throughput option to screen abiotic stress tolerance, and Ludovisi et al. (2017) have reported its successful application to phenotype the drought response in a large black poplar population consisting of 4603 individuals (503 genotypes). These examples clearly highlight how novel phenotyping technologies can widen the scale and enhance the speed of breeding programs, and need to be considered when improving novel biomass crops.
Prospects for the Improvement of Orphan Lignocellulosic Biomass Species
The tools above are effective for crop improvement, but they are available just for a handful of lignocellulosic species, and their de novo development for orphan crops would require time and adequate research investments [even if the drop of sequencing costs will soon allow association mapping and whole-genome sequencing also to novel crops (Allwright and Taylor, 2016)]. Conversely, we have seen that classical breeding alone is also time-consuming and not very effective. In this scenario, genetic tools to transfer genetic knowledge from model species to less-studied crops and to meaningfully coalesce genetic information on relevant traits across crops can be key to bridge the gap between advanced and orphan biomass species.
Translational genomics offer a possibility in this perspective, as it allows the identification of candidate genes underlying a trait of interest in a “target” organism based on its homolog(s) in a model species (Salentijn et al., 2007). Such candidate genes can then be targeted through genetic modification to obtain a desired phenotype (see section “Target traits and genetic resources to tailor novel biomass crops to MALs” for examples). This approach allowed the identification and modification of several of the candidate genes discussed in section 3, and is particularly powerful for plant clades that share high levels of genome synteny between members and include model bioenergy crops (van der Weijde et al., 2013), as grasses (Bennetzen and Freeling, 1997; Carpita and McCann, 2008). To facilitate translational genomics, several tools have been developed over the years, in the form of both genomic databases [e.g., PlantGDB2 (Dong et al., 2004), Plaza 4.0 (Van Bel et al., 2017)3, or other grass-specific databases reviewed by van der Weijde et al. (2013)] and platforms specifically designed for orphan crops lacking of a sequenced genome, but for which transcriptomes can be developed (e.g., Orphan Crops Browser; Kamei et al., 2016)4.
Most of the traits discussed in section “Target traits and genetic resources to tailor novel biomass crops to MALs” are highly quantitative, and the available knowledge on the genetics underlying them in model species is typically in the form of QTLs with no validated or known candidate genes (Barrière et al., 2007). In these cases, tools to meaningfully coalesce the information on relevant QTLs between species and make it inter-applicable in a way immediately usable in MAS or GS contexts would be very useful. Combining meta-QTL analysis approaches (Goffinet and Gerber, 2000) extended beyond species boundaries with the development of “universal markers” that are present across species but can assay intra-specific diversity for traits of interest (Ranade and Yadav, 2014) could offer promising possibilities in this direction. Such universal markers could effectively allow to project known QTLs to breeding material not included in the original panels used for QTL mapping, or even to possibly other (orphan) species, on which MAS based on universal markers could take place, without the need of de novo producing species-specific knowledge. The extensive occurrence of common genetic factors underlying complex biomass-related traits across evolutionary distant plant species (as exemplified for cell wall recalcitrance in section “Target traits and genetic resources to tailor novel biomass crops to MALs”) promises success from the application of the approach just described. However, research is needed to define to which extent common genetic determinants of traits of interest show positional conservation of their genomic organization to allow inter-species projection of QTLs and universal markers. In this direction, novel high-throughput tools to assess overall syntenic relationships between genetic elements underlying critical traits across large sets of plant genomes not even always displaying high levels of collinearity can offer promising prospects (Zhao et al., 2017; Zhao and Schranz, 2017).
Universal markers as just defined would represent a very useful tool to overcome the condition of orphan crops in which several promising species for marginal lands lay. However, these tools – as well as all the other genomic, molecular, and biotechnological approaches discussed in this review – do not represent per se a “finish line” in breeding novel promising perennial crops for marginal lands. Their effectiveness will ultimately depend by the specific ways in which breeders will integrate these tools in well-planned and modern “knowledge-based” breeding programs. Specifically, such programs will continue to largely rely on pre-breeding activities (i.e., germplasm development, dissection of the genetics underlying the traits discussed in section “Target traits and genetic resources to tailor novel biomass crops to MALs,” and development of markers), conventional crossing of promising accessions and selection within progenies, as well as ongoing schemes of population improvement through recurrent selection (especially in open-pollinated species) (Clifton-Brown et al., 2018). Nevertheless, the tools discussed in this review will allow to speed up major steps of such programs (from the genetic characterization of breeding material and the dissection of the genetic determinants of target traits, to phenotyping, marker development, or the targeting of critical genes by genetic modification), as well as precisely guide breeding activities in crops that so far have been poorly studied. This aspects will ultimately be key to ensure that novel lignocellulosic perennials will be advanced at a sufficient level for commercialization in a reasonable time, which is currently the major priority for using MALs to provide biomass for a bio-based economy.
Concluding Remarks
MALs have great potential to sustainably supply a large proportion of the biomass needed to fuel a global bio-based economy. However, the lack of crop varieties that can couple sustainability of biomass production with optimal biomass yield and quality to ensure profitable cultivation of MALs and cost-effective biomass conversion into bio-based commodities currently impedes to realize this vision. We firmly believe that plant breeding will be key to break through this impasse, and in this article we have dissected the problem of biomass provision using MALs from a plant breeding perspective. What emerges is that great progress has been made over the years in understanding the genetics underlying biomass traits. Moreover, the development of tools to study these aspects on larger scales and through quicker approaches will expand this knowledge in the future. Progress is, however, uneven among crops. While a few model species can count on an array of breeding tools and genetic knowledge to support their improvement but are unsuitable for sustainable cultivation on MALs, a wide range of locally adapted crops cannot be readily improved being paradoxically orphan in the genomic era. Therefore, our ability of creating tools to effectively transfer and coalesce the genetic knowledge on traits of interest across crops and to integrate such tools into modern, “knowledge-based” breeding programs will ultimately represent a key factor to enable the development of biomass crops tailored to the needs of MALs and to a bio-based economy.
Author Contributions
FP wrote this review, with input from LT. LT corrected the manuscript. FP and LT approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research is part of a project that has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 727698.
References
- Acharya B. S., Blanco-Canqui H. (2018). Lignocellulosic-based bioenergy and water quality parameters: a review. GCB Bioenergy 10 504–533. 10.1111/gcbb.12508 [DOI] [Google Scholar]
- Acquaah G. (2012). Breeding Cross-Pollinated Species Principles of Plant Genetics and Breeding. Hoboken, NJ: John Wiley & Sons, Ltd, 337–354. [Google Scholar]
- Acquadro A., Portis E., Lee D., Donini P., Lanteri S. (2005). Development and characterization of microsatellite markers in Cynara cardunculus L. Genome 48 217–225. 10.1139/g04-111 [DOI] [PubMed] [Google Scholar]
- Allwright M. R., Taylor G. (2016). Molecular breeding for improved second generation bioenergy crops. Trends Plant Sci. 21 43–54. 10.1016/j.tplants.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Amaducci S., Facciotto G., Bergante S., Perego A., Serra P., Ferrarini A., et al. (2017). Biomass production and energy balance of herbaceous and woody crops on marginal soils in the Po valley. GCB Bioenergy 9 31–45. 10.1111/gcbb.12341 [DOI] [Google Scholar]
- Amelework B., Shimelis H., Tongoona P., Laing M. (2015). Physiological mechanisms of drought tolerance in sorghum, genetic basis and breeding methods: a review. Afr. J. Agric. Res. 10 3029–3040. 10.5897/ajar2015.9595 [DOI] [Google Scholar]
- Ananda N., Vadlani P. V., Prasad P. V. (2011). Evaluation of drought and heat stressed grain sorghum (Sorghum bicolor) for ethanol production. Ind. Crops Prod. 33 779–782. 10.1016/j.indcrop.2011.01.007 [DOI] [Google Scholar]
- Anderson-Teixeira K. J., Davis S. C., Masters M. D., Delucia E. H. (2009). Changes in soil organic carbon under biofuel crops. GCB Bioenergy 1 75–96. 10.1111/j.1757-1707.2008.01001.x [DOI] [Google Scholar]
- Angelini L. G., Ceccarini L., O Di Nasso N. N., Bonari E. (2009). Long-term evaluation of biomass production and quality of two cardoon (Cynara cardunculus L.) cultivars for energy use. Biomass Bioenergy 33 810–816. 10.1016/j.biombioe.2008.12.004 [DOI] [Google Scholar]
- Angelini L. G., Lazzeri A., Levita G., Fontanelli D., Bozzi C. (2000). Ramie (Boehmeria nivea (L.) Gaud.) and Spanish Broom (Spartium junceum L.) fibres for composite materials: agronomical aspects, morphology and mechanical properties. Ind. Crops Prod. 11 145–161. 10.1016/s0926-6690(99)00059-x [DOI] [Google Scholar]
- Atienza S., Satovic Z., Petersen K., Dolstra O., Martin A. (2002). Preliminary genetic linkage map of Miscanthus sinensis with RAPD markers. Theor. Appl. Genet. 105 946–952. 10.1007/s00122-002-0956-7 [DOI] [PubMed] [Google Scholar]
- Atienza S. G., Satovic Z., Petersen K. K., Dolstra O., Martin A. (2003a). Identification of QTLs influencing agronomic traits in Miscanthus sinensis Anderss. I. Total height, flag-leaf height and stem diameter. Theor. Appl. Genet. 107 123–129. 10.1007/s00122-003-1220-5 [DOI] [PubMed] [Google Scholar]
- Atienza S. G., Satovic Z., Petersen K. K., Dolstra O., Martin A. (2003b). Identification of QTLs influencing combustion quality in Miscanthus sinensis Anderss. II. Chlorine and potassium content. Theor. Appl. Genet. 107 857–863. 10.1007/s00122-003-1218-z [DOI] [PubMed] [Google Scholar]
- Aylott M. J., Casella E., Tubby I., Street N., Smith P., Taylor G. (2008). Yield and spatial supply of bioenergy poplar and willow short-rotation coppice in the UK. New Phytol. 178 358–370. 10.1111/j.1469-8137.2008.02396.x [DOI] [PubMed] [Google Scholar]
- Bacci L., Baronti S., Predieri S., Di Virgilio N. (2009). Fiber yield and quality of fiber nettle (Urtica dioica L.) cultivated in Italy. Ind. Crops Prod. 29 480–484. 10.1016/j.indcrop.2008.09.005 [DOI] [Google Scholar]
- Backes A., Behr M., Xu X., Gatti E., Legay S., Predieri S., et al. (2018). Sucrose synthase gene expression analysis in the fibre nettle (Urtica dioica L.) cultivar “clone 13”. Ind. Crops Prod. 123 315–322. 10.1016/j.indcrop.2018.06.090 [DOI] [Google Scholar]
- Bahri B. A., Daverdin G., Xu X., Cheng J.-F., Barry K. W., Brummer E. C., et al. (2018). Natural variation in genes potentially involved in plant architecture and adaptation in switchgrass (Panicum virgatum L.). BMC Evol. Biol. 18:91. 10.1186/s12862-018-1193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Lee S. C., Brinton E. (2012). Waterproofing crops: effective flooding survival strategies. Plant Physiol. 160 1698–1709. 10.1104/pp.112.208173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandarra V. D. L. (2013). Efeito da aplicação de águas residuais na produtividade e na qualidade de três genótipos de Miscanthus. Costa da Caparica: Faculdade de Ciências e Tecnologia. [Google Scholar]
- Barbosa B., Costa J., Fernando A. L., Papazoglou E. G. (2015). Wastewater reuse for fiber crops cultivation as a strategy to mitigate desertification. Ind. Crops Prod. 68 17–23. 10.1016/j.indcrop.2014.07.007 [DOI] [Google Scholar]
- Barney J. N., Mann J. J., Kyser G. B., Blumwald E., Van Deynze A., Ditomaso J. M. (2009). Tolerance of switchgrass to extreme soil moisture stress: ecological implications. Plant Sci. 177 724–732. 10.1016/j.plantsci.2009.09.003 [DOI] [Google Scholar]
- Barrero R. A., Guerrero F. D., Moolhuijzen P., Goolsby J. A., Tidwell J., Bellgard S. E., et al. (2015). Shoot transcriptome of the giant reed, Arundo donax. Data Brief 3 1–6. 10.1016/j.dib.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière Y., Riboulet C., Méchin V., Maltese S., Pichon M., Cardinal A., et al. (2007). Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1 133–156. [Google Scholar]
- Bartley L., Wu Y., Zhu L., Brummer E., Saha M. (2016). Association Mapping of Cell Wall Synthesis Regulatory Genes and Cell Wall Quality in Switchgrass. Norman, OK: University of Oklahoma. [Google Scholar]
- Baute K., Van Eerd L., Robinson D., Sikkema P., Mushtaq M., Gilroyed B. (2018). Comparing the biomass yield and biogas potential of Phragmites australis with Miscanthus x giganteus and Panicum virgatum Grown in Canada. Energies 11:2198 10.3390/en11092198 [DOI] [Google Scholar]
- Bennett A. B., Isaacs R. (2014). Landscape composition influences pollinators and pollination services in perennial biofuel plantings. Agric. Ecosyst. Environ. 193 1–8. 10.1016/j.agee.2014.04.016 [DOI] [Google Scholar]
- Bennett A. B., Meehan T. D., Gratton C., Isaacs R. (2014). Modeling pollinator community response to contrasting bioenergy scenarios. PLoS One 9:e110676. 10.1371/journal.pone.0110676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Freeling M. (1997). The unified grass genome: synergy in synteny. Genome Res. 7 301–306. 10.1101/gr.7.4.301 [DOI] [PubMed] [Google Scholar]
- Bennich T., Belyazid S. (2017). The route to sustainability—Prospects and challenges of the bio-based economy. Sustainability 9:887 10.3390/su9060887 [DOI] [Google Scholar]
- Berlin S., Ghelardini L., Bonosi L., Weih M., Rönnberg-Wästljung A. C. (2014). QTL mapping of biomass and nitrogen economy traits in willows (Salix spp.) grown under contrasting water and nutrient conditions. Mol. Breed. 34 1987–2003. 10.1007/s11032-014-0157-5 [DOI] [Google Scholar]
- Bernardo R. (2016). Bandwagons I, too, have known. Theor. Appl. Genet. 129 2323–2332. 10.1007/s00122-016-2772-5 [DOI] [PubMed] [Google Scholar]
- Bhandari H., Saha M., Fasoula V., Bouton J. (2011). Estimation of genetic parameters for biomass yield in lowland switchgrass (Panicum virgatum L.). Crop Sci. 51 1525–1533. 10.2135/cropsci2010.10.0588 [DOI] [Google Scholar]
- Bhandari H., Saha M., Mascia P., Fasoula V., Bouton J. (2010). Variation among half-sib families and heritability for biomass yield and other traits in lowland switchgrass (Panicum virgatum L.). Crop Sci. 50 2355–2363. 10.2135/cropsci2010.02.0109 [DOI] [Google Scholar]
- Bita C., Gerats T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. 10.3389/fpls.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canqui H. (2016). Growing dedicated energy crops on marginal lands and ecosystem services. Soil Sci. Soc. Am. J. 80 845–858. 10.2136/sssaj2016.03.0080 [DOI] [Google Scholar]
- Blanco-Canqui H., Gilley J. E., Eisenhauer D. E., Jasa P. J., Boldt A. (2014). Soil carbon accumulation under switchgrass barriers. Agron. J. 106 2185–2192. 10.2134/agronj14.0227 [DOI] [Google Scholar]
- Bluemel M., Dally N., Jung C. (2015). Flowering time regulation in crops—what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32 121–129. 10.1016/j.copbio.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Blum A. (2011). “Drought resistance and its improvement,” in Plant Breeding for Water-Limited Environments, eds Nguyen H. T., Blum A., (New York, NY: Marcel Dekker, Inc; ), 53–152. 10.1007/978-1-4419-7491-4_3 [DOI] [Google Scholar]
- Boe A., Beck D. L. (2008). Yield components of biomass in switchgrass. Crop Sci. 48 1306–1311. 10.2135/cropsci2007.08.0482 [DOI] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. [DOI] [PubMed] [Google Scholar]
- Boléo S., Fernando A., Barbosa B., Costa J., Duarte M., Mendes B. (2015). “Remediation of soils contaminated with zinc by Miscanthus,” in WASTES 2015–Solutions, Treatments and Opportunities, eds Vilarinho C., Castro F., Russo M., (Boca Raton, FL: CRC Press; ), 37–42. [Google Scholar]
- Bonawitz N. D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44 337–363. 10.1146/annurev-genet-102209-163508 [DOI] [PubMed] [Google Scholar]
- Borkowska H., Molas R., Kupczyk A. (2009). Virginia Fanpetals (Sida hermaphrodita Rusby) Cultivated on Light Soil; Height of Yield and Biomass Productivity. Polish J. Environ. Stud. 18 563–568. [Google Scholar]
- Brachi B., Morris G. P., Borevitz J. O. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12:232. 10.1186/gb-2011-12-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H., Stettler R. F. (1995). Molecular genetics of growth and development in populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. . Genetics 139 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H., Villar M., Watson B., Otto K., Stewart S., Stettler R. (1994). Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor. Appl. Genet. 89 167–178. 10.1007/bf00225137 [DOI] [PubMed] [Google Scholar]
- Braun D. M., Wang L., Ruan Y.-L. (2013). Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 65 1713–1735. 10.1093/jxb/ert416 [DOI] [PubMed] [Google Scholar]
- Brereton N. J., Pitre F. E., Hanley S. J., Ray M. J., Karp A., Murphy R. J. (2010). QTL mapping of enzymatic saccharification in short rotation coppice willow and its independence from biomass yield. Bioenergy Res. 3 251–261. 10.1007/s12155-010-9077-3 [DOI] [Google Scholar]
- Brondani R. P., Williams E. R., Brondani C., Grattapaglia D. (2006). A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus. BMC Plant Biol. 6:20. 10.1186/1471-2229-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., Browne C., et al. (2009). The genetic architecture of maize flowering time. Science 325 714–718. [DOI] [PubMed] [Google Scholar]
- Cai X., Zhang X., Wang D. (2010). Land availability for biofuel production. Environ. Sci. Technol. 45 334–339. 10.1021/es103338e [DOI] [PubMed] [Google Scholar]
- Campbell J. E., Lobell D. B., Genova R. C., Field C. B. (2008). The global potential of bioenergy on abandoned agriculture lands. Environ. Sci. Technol. 42 5791–5794. 10.1021/es800052w [DOI] [PubMed] [Google Scholar]
- Canavan K., Paterson I. D., Hill M. P. (2017). Exploring the origin and genetic diversity of the giant reed, Arundo donax in South Africa. Invasive Plant Sci. Manage. 10 53–60. 10.1017/inp.2016.5 [DOI] [Google Scholar]
- Carlsson G., Mårtensson L. M., Prade T., Svensson S. E., Jensen E. S. (2017). Perennial species mixtures for multifunctional production of biomass on marginal land. GCB Bioenergy 9 191–201. 10.1111/gcbb.12373 [DOI] [Google Scholar]
- Carocha V., Soler M., Hefer C., Cassan-Wang H., Fevereiro P., Myburg A. A., et al. (2015). Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol. 206 1297–1313. 10.1111/nph.13313 [DOI] [PubMed] [Google Scholar]
- Carpita N. C., McCann M. C. (2008). Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci. 13 415–420. 10.1016/j.tplants.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Carroll A., Somerville C. (2009). Cellulosic biofuels. Annu. Rev. Plant Biol. 60 165–182. [DOI] [PubMed] [Google Scholar]
- Casler M. D., Tobias C. M., Kaeppler S. M., Buell C. R., Wang Z.-Y., Cao P., et al. (2011). The switchgrass genome: tools and strategies. Plant Genome 4 273–282. 10.3835/plantgenome2011.10.0026 [DOI] [Google Scholar]
- Cervera M.-T., Storme V., Ivens B., Gusmao J., Liu B. H., Hostyn V., et al. (2001). Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158 787–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvat M., Perez G., Hedde M., Lamy I. (2014). Establishment of bioenergy crops on metal contaminated soils stimulates belowground fauna. Biomass Bioenergy 62 207–211. 10.1016/j.biombioe.2014.01.042 [DOI] [Google Scholar]
- Chen C.-L. (2018). Genetic Diversity and Mechanisms of Salt Tolerance of Miscanthus. Ph.D. thesis, Wageningen University, Wageningen. [Google Scholar]
- Chen F., Dixon R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25 759–761. 10.1038/nbt1316 [DOI] [PubMed] [Google Scholar]
- Chimento C., Almagro M., Amaducci S. (2016). Carbon sequestration potential in perennial bioenergy crops: the importance of organic matter inputs and its physical protection. GCB Bioenergy 8 111–121. 10.1111/gcbb.12232 [DOI] [Google Scholar]
- Clifton-Brown J., Harfouche A., Casler M. D., Dylan Jones H., Macalpine W. J., Murphy-Bokern D., et al. (2018). Breeding progress and preparedness for mass-scale deployment of perennial lignocellulosic biomass crops switchgrass, miscanthus, willow and poplar. GCB Bioenergy 11 118–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton-Brown J., Jones M. (1997). The thermal response of leaf extension rate in genotypes of the C4–grass Miscanthus: an important factor in determining the potential productivity of different genotypes. J. Exp. Bot. 48 1573–1581. 10.1093/jexbot/48.313.1573 [DOI] [Google Scholar]
- Clifton-Brown J., Lewandowski I. (2000). Overwintering problems of newly established Miscanthus plantations can be overcome by identifying genotypes with improved rhizome cold tolerance. New Phytol. 148 287–294. 10.1046/j.1469-8137.2000.00764.x [DOI] [Google Scholar]
- Cosentino S. L., Copani V., D’agosta G. M., Sanzone E., Mantineo M. (2006). First results on evaluation of Arundo donax L. clones collected in Southern Italy. Ind. Crops Prod. 23 212–222. 10.1016/j.indcrop.2005.06.004 [DOI] [Google Scholar]
- Cosentino S. L., Copani V., Scalici G., Scordia D., Testa G. (2015). Soil erosion mitigation by perennial species under Mediterranean environment. Bioenergy Res. 8 1538–1547. 10.1007/s12155-015-9690-2 [DOI] [Google Scholar]
- Cosgrove D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- Costa J., Barbosa B., Fernando A. L. (2016). “Wastewaters reuse for energy crops cultivation,” in Proceedings of the Doctoral Conference on Computing, Electrical and Industrial Systems, Costa de Caparica, 507–514. 10.1007/978-3-319-31165-4_47 [DOI] [Google Scholar]
- Curt M. D., Fernández J. (1990). Production of Nicotiana glauca RC Graham aerial biomass in relation to irrigation regime. Biomass 23 103–115. 10.1016/0144-4565(90)90029-j [DOI] [Google Scholar]
- da Costa R. M., Pattathil S., Avci U., Winters A., Hahn M. G., Bosch M. (2019). Desirable plant cell wall traits for higher-quality miscanthus lignocellulosic biomass. Biotechnol. Biofuels 12:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Hu Q., Cai Q., Feng K., Ye N., Tuskan G. A., et al. (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res. 24 1274–1277. 10.1038/cr.2014.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily G. C. (1995). Restoring value to the world’s degraded lands. Science 269 350–354. 10.1126/science.269.5222.350 [DOI] [PubMed] [Google Scholar]
- Dale V. H., Kline K. L., Wiens J., Fargione J. (2010). Biofuels: Implications for Land Use and Biodiversity. Washington, DC: Ecological Society of America. [Google Scholar]
- Das M. K., Fuentes R. G., Taliaferro C. M. (2004). Genetic variability and trait relationships in switchgrass. Crop Sci. 44 443–448. 10.2135/cropsci2004.4430 [DOI] [Google Scholar]
- Dauber J., Brown C., Fernando A. L., Finnan J., Krasuska E., Ponitka J., et al. (2012). Bioenergy from “surplus” land: environmental and socio-economic implications. BioRisk 7 5–50. 10.3897/biorisk.7.3036 [DOI] [Google Scholar]
- Dauber J., Jones M. B., Stout J. C. (2010). The impact of biomass crop cultivation on temperate biodiversity. GCB Bioenergy 2 289–309. 10.1111/j.1757-1707.2010.01058.x [DOI] [Google Scholar]
- Davey C. L., Robson P., Hawkins S., Farrar K., Clifton-Brown J. C., Donnison I. S., et al. (2017). Genetic relationships between spring emergence, canopy phenology, and biomass yield increase the accuracy of genomic prediction in Miscanthus. J. Exp. Bot. 68 5093–5102. 10.1093/jxb/erx339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. C., Dohleman F. G., Long S. P. (2011). The global potential for Agave as a biofuel feedstock. GCB Bioenergy 3 68–78. 10.1111/j.1757-1707.2010.01077.x [DOI] [Google Scholar]
- De Oliveira Silva F. M., Lichtenstein G., Alseekh S., Rosado-Souza L., Conte M., Suguiyama V. F., et al. (2018). The genetic architecture of photosynthesis and plant growth-related traits in tomato. Plant Cell Environ. 41 327–341. 10.1111/pce.13084 [DOI] [PubMed] [Google Scholar]
- de Vries S. C., Van De Ven G. W., Van Ittersum M. K., Giller K. E. (2010). Resource use efficiency and environmental performance of nine major biofuel crops, processed by first-generation conversion techniques. Biomass Bioenergy 34 588–601. 10.1016/j.biombioe.2010.01.001 [DOI] [Google Scholar]
- Di Michele M., Chiatante D., Plomion C., Scippa G. S. (2006). A proteomic analysis of Spanish broom (Spartium junceum L.) root growing on a slope condition. Plant Sci. 170 926–935. 10.1016/j.plantsci.2005.12.020 [DOI] [Google Scholar]
- Di Nasso N. N., Lasorella M., Roncucci N., Bonari E. (2015). Soil texture and crop management affect switchgrass (Panicum virgatum L.) productivity in the Mediterranean. Ind. Crops Prod. 65 21–26. 10.1016/j.indcrop.2014.11.017 [DOI] [Google Scholar]
- Dietz T., Börner J., Förster J., Von Braun J. (2018). Governance of the bioeconomy: a global comparative study of national bioeconomy strategies. Sustainability 10:3190 10.3390/su10093190 [DOI] [Google Scholar]
- Doblin M. S., Pettolino F. A., Wilson S. M., Campbell R., Burton R. A., Fincher G. B., et al. (2009). A barley cellulose synthase-like CSLH gene mediates (1, 3; 1, 4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 5996–6001. 10.1073/pnas.0902019106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohleman F. G., Long S. P. (2009). More productive than maize in the Midwest: how does Miscanthus do it? Plant Physiol. 150 2104–2115. 10.1104/pp.109.139162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Thames S., Liu L., Smith M. W., Yan L., Wu Y. (2015). QTL mapping for reproductive maturity in lowland switchgrass populations. Bioenergy Res. 8 1925–1937. 10.1007/s12155-015-9651-9 [DOI] [Google Scholar]
- Dong Q., Schlueter S. D., Brendel V. (2004). PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 32 D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Lu W., Quan M., Xiao L., Song F., Li P., et al. (2018). Genome-wide association studies to improve wood properties: challenges and prospects. Front. Plant Sci. 9:1912. 10.3389/fpls.2018.01912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J. G., Strabala T. J., Wagner A. (2013). Potential transgenic routes to increase tree biomass. Plant Sci. 212 72–101. 10.1016/j.plantsci.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Dwiyanti M. S., Stewart J. R., Yamada T. (2014). Forages for feedstocks of biorefineries in temperate environments: review of lignin research in bioenergy crops and some insight into Miscanthus studies. Crop Pasture Sci. 65 1199–1206. [Google Scholar]
- Emerson R., Hoover A., Ray A., Lacey J., Cortez M., Payne C., et al. (2014). Drought effects on composition and yield for corn stover, mixed grasses, and Miscanthus as bioenergy feedstocks. Biofuels 5 275–291. 10.1080/17597269.2014.913904 [DOI] [Google Scholar]
- Evangelistella C., Valentini A., Ludovisi R., Firrincieli A., Fabbrini F., Scalabrin S., et al. (2017). De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock. Biotechnol. Biofuels 10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J., Sanciangco M. D., Lau K. H., Crisovan E., Barry K., Daum C., et al. (2017). Extensive genetic diversity is present within North American switchgrass germplasm. Plant Genome 11:170055. 10.3835/plantgenome2017.06.0055 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog A. M., Neves L. G., Resende M. F., Vazquez A. I., Campos G., Dervinis C., et al. (2017). Genome-wide association study reveals putative regulators of bioenergy traits in Populus deltoides. New Phytol. 213 799–811. 10.1111/nph.14154 [DOI] [PubMed] [Google Scholar]
- Fargione J., Hill J., Tilman D., Polasky S., Hawthorne P. (2008). Land clearing and the biofuel carbon debt. Science 319 1235–1238. 10.1126/science.1152747 [DOI] [PubMed] [Google Scholar]
- Farrell A. D., Clifton-Brown J., Lewandowski I., Jones M. (2006). Genotypic variation in cold tolerance influences the yield of Miscanthus. Ann. Appl. Biol. 149 337–345. 10.1111/j.1744-7348.2006.00099.x [DOI] [Google Scholar]
- Farrell A. E., Plevin R. J., Turner B. T., Jones A. D., O’hare M., Kammen D. M. (2006). Ethanol can contribute to energy and environmental goals. Science 311 506–508. 10.1126/science.1121416 [DOI] [PubMed] [Google Scholar]
- Fernandez M. G. S., Bao Y., Tang L., Schnable P. S. (2017). A high-throughput, field-based phenotyping technology for tall biomass crops. Plant Physiol. 174 2008–2022. 10.1104/pp.17.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M. G. S., Becraft P. W., Yin Y., Lübberstedt T. (2009). From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci. 14 454–461. 10.1016/j.tplants.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Fernando A. L., Costa J., Barbosa B., Monti A., Rettenmaier N. (2018). Environmental impact assessment of perennial crops cultivation on marginal soils in the Mediterranean Region. Biomass Bioenergy 111 174–186. 10.1016/j.biombioe.2017.04.005 [DOI] [Google Scholar]
- Ferreira S., Hjernø K., Larsen M., Wingsle G., Larsen P., Fey S., et al. (2006). Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann. Bot. 98 361–377. 10.1093/aob/mcl106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C. B., Campbell J. E., Lobell D. B. (2008). Biomass energy: the scale of the potential resource. Trends Ecol. Evol. 23 65–72. 10.1016/j.tree.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Flood P. J., Harbinson J., Aarts M. G. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16 327–335. 10.1016/j.tplants.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Francaviglia R., Bruno A., Falcucci M., Farina R., Renzi G., Russo D. E., et al. (2016). Yields and quality of Cynara cardunculus L. wild and cultivated cardoon genotypes. A case study from a marginal land in Central Italy. Eur. J. Agron. 72 10–19. 10.1016/j.eja.2015.09.014 [DOI] [Google Scholar]
- Franzaring J., Schmid I., Bäuerle L., Gensheimer G., Fangmeier A. (2014). Investigations on plant functional traits, epidermal structures and the ecophysiology of the novel bioenergy species Sida hermaphrodita Rusby and Silphium perfoliatum L. J. Appl. Bot. Food Qual. 87 36–45. [Google Scholar]
- Freeman J. S., Whittock S. P., Potts B. M., Vaillancourt R. E. (2009). QTL influencing growth and wood properties in Eucalyptus globulus. Tree Genet. Genomes 5 713–722. 10.1007/s11295-009-0222-0 [DOI] [Google Scholar]
- Fritsche U. R., Sims R. E., Monti A. (2010). Direct and indirect land-use competition issues for energy crops and their sustainable production–an overview. Biofuel. Bioprod. Biorefin. 4 692–704. 10.1002/bbb.258 [DOI] [Google Scholar]
- Fu C., Mielenz J. R., Xiao X., Ge Y., Hamilton C. Y., Rodriguez M., et al. (2011). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108 3803–3808. 10.1073/pnas.1100310108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielle B., Bamière L., Caldes N., De Cara S., Decocq G., Ferchaud F., et al. (2014). Paving the way for sustainable bioenergy in Europe: technological options and research avenues for large-scale biomass feedstock supply. Renew. Sustain. Energy Rev. 33 11–25. 10.1016/j.rser.2014.01.050 [DOI] [Google Scholar]
- Gardiner M. A., Tuell J. K., Isaacs R., Gibbs J., Ascher J. S., Landis D. A. (2010). Implications of three biofuel crops for beneficial arthropods in agricultural landscapes. Bioenergy Res. 3 6–19. 10.1007/s12155-009-9065-7 [DOI] [Google Scholar]
- Gaudet M., Jorge V., Paolucci I., Beritognolo I., Mugnozza G. S., Sabatti M. (2008). Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet. Genomes 4 25–36. 10.1007/s11295-007-0085-1 [DOI] [Google Scholar]
- Ge C., Ai X., Jia S., Yang Y., Che L., Yi Z., et al. (2018). Interspecific genetic maps in Miscanthus floridulus and M. sacchariflorus accelerate detection of QTLs associated with plant height and inflorescence. Mol. Genet. Genomics 294 35–45. 10.1007/s00438-018-1486-6 [DOI] [PubMed] [Google Scholar]
- Gelfand I., Sahajpal R., Zhang X., Izaurralde R. C., Gross K. L., Robertson G. P. (2013). Sustainable bioenergy production from marginal lands in the US Midwest. Nature 493 514–517. 10.1038/nature11811 [DOI] [PubMed] [Google Scholar]
- Gelfand I., Zenone T., Jasrotia P., Chen J., Hamilton S. K., Robertson G. P. (2011). Carbon debt of Conservation Reserve Program (CRP) grasslands converted to bioenergy production. Proc. Natl. Acad. Sci. U.S.A. 108 13864–13869. 10.1073/pnas.1017277108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer W., Argent R., Walawender W. (2007). Biomass properties and gasification behavior of 7-year-old Siberian elm. Wood Fiber Sci. 19 176–182. [Google Scholar]
- Ghannoum O., Evans J., Von Caemmerer S. (2011). “Nitrogen and water use efficiency of C4 plants,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds Raghavendra A. S., Sage R. F., (Berlin: Springer; ). [Google Scholar]
- Gibbs H. K., Johnston M., Foley J. A., Holloway T., Monfreda C., Ramankutty N., et al. (2008). Carbon payback times for crop-based biofuel expansion in the tropics: the effects of changing yield and technology. Environ. Res. Lett. 3:034001 10.1088/1748-9326/3/3/034001 [DOI] [Google Scholar]
- Gifford J. M., Chae W. B., Swaminathan K., Moose S. P., Juvik J. A. (2015). Mapping the genome of Miscanthus sinensis for QTL associated with biomass productivity. GCB Bioenergy 7 797–810. 10.1111/gcbb.12201 [DOI] [Google Scholar]
- Glass M., Barkwill S., Unda F., Mansfield S. D. (2015). Endo-β-1, 4-glucanases impact plant cell wall development by influencing cellulose crystallization. J. Integr. Plant Biol. 57 396–410. 10.1111/jipb.12353 [DOI] [PubMed] [Google Scholar]
- Glover J. D., Culman S. W., Dupont S. T., Broussard W., Young L., Mangan M. E., et al. (2010). Harvested perennial grasslands provide ecological benchmarks for agricultural sustainability. Agric. Ecosyst. Environ. 137 3–12. 10.1016/j.agee.2009.11.001 [DOI] [Google Scholar]
- Goffinet B., Gerber S. (2000). Quantitative trait loci: a meta-analysis. Genetics 155 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D., Bertolucci F., Sederoff R. (1995). Genetic mapping of QTLs controlling vegetative propagation in Eucalyptus grandis and E. urophylla using a pseudo-testcross strategy and RAPD markers. Theor. Appl. Genet. 90 933–947. 10.1007/bf00222906 [DOI] [PubMed] [Google Scholar]
- Grattapaglia D., Bertolucci F. L., Penchel R., Sederoff R. R. (1996). Genetic mapping of quantitative trait loci controlling growth and wood quality traits in Eucalyptus grandis using a maternal half-sib family and RAPD markers. Genetics 144 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D., Kirst M. (2008). Eucalyptus applied genomics: from gene sequences to breeding tools. New Phytol. 179 911–929. 10.1111/j.1469-8137.2008.02503.x [DOI] [PubMed] [Google Scholar]
- Grattapaglia D., Sederoff R. (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen P. L., Culetic A., Boschian L., Krupinska K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82 603–622. 10.1007/s11103-013-0013-8 [DOI] [PubMed] [Google Scholar]
- Gross S. M., Martin J. A., Simpson J., Abraham-Juarez M. J., Wang Z., Visel A. (2013). De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics 14:563. 10.1186/1471-2164-14-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberl H., Beringer T., Bhattacharya S. C., Erb K.-H., Hoogwijk M. (2010). The global technical potential of bio-energy in 2050 considering sustainability constraints. Curr. Opin. Environ. Sustain. 2 394–403. 10.1016/j.cosust.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberl H., Erb K.-H., Krausmann F., Bondeau A., Lauk C., Müller C., et al. (2011). Global bioenergy potentials from agricultural land in 2050: sensitivity to climate change, diets and yields. Biomass Bioenergy 35 4753–4769. 10.1016/j.biombioe.2011.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale A. L., Viator R. P., Veremis J. C. (2014). Identification of freeze tolerant Saccharum spontaneum accessions through a pot-based study for use in sugarcane germplasm enhancement for adaptation to temperate climates. Biomass Bioenergy 61 53–57. 10.1016/j.biombioe.2013.11.015 [DOI] [Google Scholar]
- Hamilton S., Hussain M., Bhardwaj A., Basso B., Robertson G. (2015). Comparative water use by maize, perennial crops, restored prairie, and poplar trees in the US Midwest. Environ. Res. Lett. 10:064015 10.1088/1748-9326/10/6/064015 [DOI] [Google Scholar]
- Hanley S., Barker J., Van Ooijen J., Aldam C., Harris S., Åhman I., et al. (2002). A genetic linkage map of willow (Salix viminalis) based on AFLP and microsatellite markers. Theor. Appl. Genet. 105 1087–1096. 10.1007/s00122-002-0979-0 [DOI] [PubMed] [Google Scholar]
- Hanley S., Mallott M., Karp A. (2006). Alignment of a Salix linkage map to the Populus genomic sequence reveals macrosynteny between willow and poplar genomes. Tree Genet. Genomes 3 35–48. 10.1007/s11295-006-0049-x [DOI] [Google Scholar]
- Hansson P.-A., Fredriksson H. (2004). Use of summer harvested common reed (Phragmites australis) as nutrient source for organic crop production in Sweden. Agric. Ecosyst. Environ. 102 365–375. 10.1016/j.agee.2003.08.005 [DOI] [Google Scholar]
- Harris D., Debolt S. (2008). Relative crystallinity of plant biomass: studies on assembly, adaptation and acclimation. PLoS One 3:e2897. 10.1371/journal.pone.0002897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D., Stork J., Debolt S. (2009). Genetic modification in cellulose-synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. GCB Bioenergy 1 51–61. 10.1111/j.1757-1707.2009.01000.x [DOI] [Google Scholar]
- Hartl A., Vogl C. R. (2002). Dry matter and fiber yields, and the fiber characteristics of five nettle clones (Urtica dioica L.) organically grown in Austria for potential textile use. Am. J. Altern. Agric. 17 195–200. 10.1079/aja2002195 [DOI] [Google Scholar]
- Haughton A. J., Bohan D. A., Clark S. J., Mallott M. D., Mallott V., Sage R., et al. (2016). Dedicated biomass crops can enhance biodiversity in the arable landscape. GCB Bioenergy 8 1071–1081. 10.1111/gcbb.12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner E. L., Sorrells M. E., Jannink J.-L. (2009). Genomic selection for crop improvement. Crop Sci. 49 1–12. 10.2135/cropsci2008.08.0512 [DOI] [Google Scholar]
- Hikosaka K. (2010). Mechanisms underlying interspecific variation in photosynthetic capacity across wild plant species. Plant Biotechnol. 27 223–229. 10.5511/plantbiotechnology.27.223 [DOI] [Google Scholar]
- Hilley J., Truong S., Olson S., Morishige D., Mullet J. (2016). Identification of Dw1, a regulator of sorghum stem internode length. PLoS One 11:e0151271. 10.1371/journal.pone.0151271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchee M., Zhang C., Chang S., Cunningham M., Hammond W., Nehra N. (2011). Biotech Eucalyptus can sustainably address society’s need for wood: the example of freeze tolerant Eucalyptusin the southeastern U.S. BMC Proc. 5:I24 10.1186/1753-6561-5-S7-I24 [DOI] [Google Scholar]
- Hisano H., Nandakumar R., Wang Z.-Y. (2011). “Genetic modification of lignin biosynthesis for improved biofuel production,” inBiofuels, eds Tomes D., Lakshmanan P., Songstad D., (New York, NY: Springer; ), 223–235. 10.1007/978-1-4419-7145-6_12 [DOI] [Google Scholar]
- Hollender C. A., Dardick C. (2015). Molecular basis of angiosperm tree architecture. New Phytol. 206 541–556. 10.1111/nph.13204 [DOI] [PubMed] [Google Scholar]
- Hoogwijk M., Faaij A., Van Den Broek R., Berndes G., Gielen D., Turkenburg W. (2003). Exploration of the ranges of the global potential of biomass for energy. Biomass Bioenergy 25 119–133. 10.1016/s0961-9534(02)00191-5 [DOI] [Google Scholar]
- Houston K., Tucker M. R., Chowdhury J., Shirley N., Little A. (2016). The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7:984. 10.3389/fpls.2016.00984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isikgor F. H., Becer C. R. (2015). Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 6 4497–4559. 10.1039/c5py00263j [DOI] [Google Scholar]
- Jensen E., Farrar K., Thomas-Jones S., Hastings A., Donnison I., Clifton-Brown J. (2011). Characterization of flowering time diversity in Miscanthus species. GCB Bioenergy 3 387–400. 10.1111/j.1757-1707.2011.01097.x [DOI] [Google Scholar]
- Jones M. B., Finnan J., Hodkinson T. R. (2015). Morphological and physiological traits for higher biomass production in perennial rhizomatous grasses grown on marginal land. GCB Bioenergy 7 375–385. 10.1111/gcbb.12203 [DOI] [Google Scholar]
- Jones R., Le-Bas C., Nachtergaele F., Rossiter D., Van J., Orshoven R. S., et al. (2014). Updated common bio-physical criteria to define natural constraints for agriculture in Europe. Definition and scientific justification for the common criteria. JRC Science and Policy Reports EUR 26638 EN, 68, Ispra 10.2788/79958 [DOI] [Google Scholar]
- Joshi C. P., Thammannagowda S., Fujino T., Gou J.-Q., Avci U., Haigler C. H., et al. (2011). Perturbation of wood cellulose synthesis causes pleiotropic effects in transgenic aspen. Mol. Plant 4 331–345. 10.1093/mp/ssq081 [DOI] [PubMed] [Google Scholar]
- Kamei C. L. A., Severing E. I., Dechesne A., Furrer H., Dolstra O., Trindade L. M. (2016). Orphan crops browser: a bridge between model and orphan crops. Mol. Breed. 36:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp A., Shield I. (2008). Bioenergy from plants and the sustainable yield challenge. New Phytol. 179 15–32. 10.1111/j.1469-8137.2008.02432.x [DOI] [PubMed] [Google Scholar]
- Kendurkar S. V., Rangaswamy M. (2018). “Genetic Transformation in Eucalyptus,” inBiotechnologies of Crop Improvement, Vol. 2 eds Gosal S. S, Wani S. H., (Berlin: Springer; ), 335–366. 10.1007/978-3-319-90650-8_13 [DOI] [Google Scholar]
- Kenney W., Sennerby-Forsse L., Layton P. (1990). A review of biomass quality research relevant to the use of poplar and willow for energy conversion. Biomass 21 163–188. 10.1016/0144-4565(90)90063-p [DOI] [Google Scholar]
- Khudamrongsawat J., Tayyar R., Holt J. S. (2004). Genetic diversity of giant reed (Arundo donax) in the Santa Ana River, California. Weed Sci. 52 395–405. 10.1614/ws-03-120r1 [DOI] [Google Scholar]
- Kim C., Zhang D., Auckland S. A., Rainville L. K., Jakob K., Kronmiller B., et al. (2012). SSR-based genetic maps of Miscanthus sinensis and M. sacchariflorus, and their comparison to sorghum. Theor. Appl. Genet. 124 1325–1338. 10.1007/s00122-012-1790-1 [DOI] [PubMed] [Google Scholar]
- Kirst M., Myburg A. A., De León J. P., Kirst M. E., Scott J., Sederoff R. (2004). Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of eucalyptus. Plant Physiol. 135 2368–2378. 10.1104/pp.103.037960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Kim C., Goff V. H., Zhang D., Paterson A. H. (2015). Genetic analysis of rhizomatousness and its relationship with vegetative branching of recombinant inbred lines of Sorghum bicolor × S. propinquum. Am. J. Bot. 102 718–724. 10.3732/ajb.1500035 [DOI] [PubMed] [Google Scholar]
- Kuehl Y. (2015). “Resources, yield, and volume of bamboos,” in Bamboo, eds Köhl W., Liese M., (Basel: Springer; ), 91–111. 10.1007/978-3-319-14133-6_4 [DOI] [Google Scholar]
- Lamers J. P. A., Khamzina A. (2008). Fuelwood production in the degraded agricultural areas of the Aral Sea Basin, Uzbekistan. Bois For. Trop. 297 47–57. [Google Scholar]
- Landis D. A., Gratton C., Jackson R. D., Gross K. L., Duncan D. S., Liang C., et al. (2018). Biomass and biofuel crop effects on biodiversity and ecosystem services in the North Central US. Biomass Bioenergy 114 18–29. 10.1016/j.biombioe.2017.02.003 [DOI] [Google Scholar]
- Lanteri S., Acquadro A., Comino C., Mauro R., Mauromicale G., Portis E. (2006). A first linkage map of globe artichoke (Cynara cardunculus var. scolymus L.) based on AFLP, S-SAP, M-AFLP and microsatellite markers. Theor. Appl. Genet. 112 1532–1542. 10.1007/s00122-006-0256-8 [DOI] [PubMed] [Google Scholar]
- Lanteri S., Portis E., Acquadro A., Mauro R. P., Mauromicale G. (2012). Morphology and SSR fingerprinting of newly developed Cynara cardunculus genotypes exploitable as ornamentals. Euphytica 184 311–321. 10.1007/s10681-011-0509-8 [DOI] [Google Scholar]
- Lauria M., Molinari F., Motto M. (2015). “Genetic strategies to enhance plant biomass yield and quality-related traits for bio-renewable fuel and chemical productions,” in Plants for the Future, ed. El-Shemy H., (Rijeka: InTech; ). [Google Scholar]
- Lawson T., Kramer D. M., Raines C. A. (2012). Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr. Opin. Biotechnol. 23 215–220. 10.1016/j.copbio.2011.12.012 [DOI] [PubMed] [Google Scholar]
- Le Gall H., Philippe F., Domon J.-M., Gillet F., Pelloux J., Rayon C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4 112–166. 10.3390/plants4010112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDuc S. D., Zhang X., Clark C. M., Izaurralde R. C. (2017). Cellulosic feedstock production on Conservation Reserve Program land: potential yields and environmental effects. Gcb Bioenergy 9 460–468. 10.1111/gcbb.12352 [DOI] [Google Scholar]
- Levine U. Y., Teal T. K., Robertson G. P., Schmidt T. M. (2011). Agriculture’s impact on microbial diversity and associated fluxes of carbon dioxide and methane. ISME J. 5 1683–1691. 10.1038/ismej.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski I. (2016). “The role of perennial biomass crops in a growing bioeconomy,” in Perennial Biomass Crops for a Resource-Constrained World, eds Barth S., Murphy-Bokern D., Kalinina O., Taylor G., Jones M. (Basel: Springer; ), 3–13. 10.1007/978-3-319-44530-4_1 [DOI] [Google Scholar]
- Li M., Pu Y., Ragauskas A. J. (2016). Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 4:45 10.3389/fchem.2016.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere H., Kim T. N., Werling B. P., Meehan T. D., Landis D. A., Gratton C. (2015). Trophic cascades in agricultural landscapes: indirect effects of landscape composition on crop yield. Ecol. Appl. 25 652–661. 10.1890/14-0570.1 [DOI] [PubMed] [Google Scholar]
- Lin Y.-R., Schertz K. F., Paterson A. H. (1995). Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141 391–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Luo L., Zheng L. (2018). Lignins: biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19:E335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Clark L. V., Swaminathan K., Gifford J. M., Juvik J. A., Sacks E. J. (2016). High-density genetic map of Miscanthus sinensis reveals inheritance of zebra stripe. GCB Bioenergy 8 616–630. 10.1111/gcbb.12275 [DOI] [Google Scholar]
- Liu W., Mi J., Song Z., Yan J., Li J., Sang T. (2014). Long-term water balance and sustainable production of Miscanthus energy crops in the Loess Plateau of China. Biomass Bioenergy 62 47–57. 10.1016/j.biombioe.2014.01.018 [DOI] [Google Scholar]
- Liu Y., Zhang X., Miao J., Huang L., Frazier T., Zhao B. (2014). Evaluation of salinity tolerance and genetic diversity of thirty-three switchgrass (Panicum virgatum) populations. Bioenergy Res. 7 1329–1342. 10.1007/s12155-014-9466-0 [DOI] [Google Scholar]
- Loque D., Scheller H. V., Pauly M. (2015). Engineering of plant cell walls for enhanced biofuel production. Curr. Opin. Plant Biol. 25 151–161. 10.1016/j.pbi.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Lord R. (2015). Reed canarygrass (Phalaris arundinacea) outperforms Miscanthus or willow on marginal soils, brownfield and non-agricultural sites for local, sustainable energy crop production. Biomass Bioenergy 78 110–125. 10.1016/j.biombioe.2015.04.015 [DOI] [Google Scholar]
- Lowry D. B., Taylor S. H., Bonnette J., Aspinwall M. J., Asmus A. L., Keitt T. H., et al. (2015). QTLs for biomass and developmental traits in switchgrass (Panicum virgatum). Bioenergy Res. 8 1856–1867. 10.1007/s12155-015-9629-7 [DOI] [Google Scholar]
- Lu F., Lipka A. E., Glaubitz J., Elshire R., Cherney J. H., Casler M. D., et al. (2013). Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 9:e1003215. 10.1371/journal.pgen.1003215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübberstedt T., Melchinger A. E., Schön C. C., Utz H. F., Klein D. (1997). QTL mapping in testcrosses of European flint lines of maize: I. Comparison of different testers for forage yield traits. Crop Sci. 37 921–931. 10.2135/cropsci1997.0011183x003700030037x [DOI] [Google Scholar]
- Ludovisi R., Tauro F., Salvati R., Khoury S., Mugnozza Scarascia G., Harfouche A. (2017). UAV-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front. Plant Sci. 8:1681. 10.3389/fpls.2017.01681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.-F., Jensen E., Alexandrov N., Troukhan M., Zhang L., Thomas-Jones S., et al. (2012). High resolution genetic mapping by genome sequencing reveals genome duplication and tetraploid genetic structure of the diploid Miscanthus sinensis. PLoS One 7:e33821. 10.1371/journal.pone.0033821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J. M., Virtue J. G., Williams C., Preston C. (2017). Genetic diversity of giant reed (A rundo donax) in Australia. Weed Biol. Manage. 17 17–28. 10.1111/wbm.12111 [DOI] [Google Scholar]
- Mandrou E., Denis M., Plomion C., Salin F., Mortier F., Gion J.-M. (2014). Nucleotide diversity in lignification genes and QTNs for lignin quality in a multi-parental population of Eucalyptus urophylla. Tree Genet. Genomes 10 1281–1290. 10.1007/s11295-014-0760-y [DOI] [Google Scholar]
- Marques C., Araujo J., Ferreira J., Whetten R., O’malley D., Liu B.-H., et al. (1998). AFLP genetic maps of Eucalyptus globulus and E. tereticornis. Theor. Appl. Genet. 96 727–737. 10.1007/s001220050795 [DOI] [Google Scholar]
- Marques C., Brondani R., Grattapaglia D., Sederoff R. (2002). Conservation and synteny of SSR loci and QTLs for vegetative propagation in four Eucalyptus species. Theor. Appl. Genet. 105 474–478. 10.1007/s00122-002-0899-z [DOI] [PubMed] [Google Scholar]
- Marron N., Bastien C., Sabatti M., Taylor G., Ceulemans R. (2006). Plasticity of growth and sylleptic branchiness in two poplar families grown at three sites across Europe. Tree Physiol. 26 935–946. 10.1093/treephys/26.7.935 [DOI] [PubMed] [Google Scholar]
- Martin E., Cravero V., Cointry E. (2015). “Quantitative trait loci (QTLs) related to biomass production in Cynara cardunculus L,” in Proceedings of the IX International Symposium on Artichoke, Cardoon and Their Wild Relatives, La Plata, 189–196. 10.17660/actahortic.2016.1147.28 [DOI] [Google Scholar]
- Mauromicale G., Sortino O., Pesce G. R., Agnello M., Mauro R. P. (2014). Suitability of cultivated and wild cardoon as a sustainable bioenergy crop for low input cultivation in low quality Mediterranean soils. Ind. Crops Prod. 57 82–89. 10.1016/j.indcrop.2014.03.013 [DOI] [Google Scholar]
- McCormick K., Kautto N. (2013). The bioeconomy in Europe: an overview. Sustainability 5 2589–2608. 10.3390/su5062589 [DOI] [Google Scholar]
- McIsaac G. F., David M. B., Mitchell C. A. (2010). Miscanthus and switchgrass production in central Illinois: impacts on hydrology and inorganic nitrogen leaching. J. Environ. Qual. 39 1790–1799. 10.2134/jeq2009.0497 [DOI] [PubMed] [Google Scholar]
- McKown A. D., Klápštì J., Guy R. D., Geraldes A., Porth I., Hannemann J., et al. (2014). Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytol. 203 535–553. 10.1111/nph.12815 [DOI] [PubMed] [Google Scholar]
- Meehan T. D., Hurlbert A. H., Gratton C. (2010). Bird communities in future bioenergy landscapes of the Upper Midwest. Proc. Natl. Acad. Sci. U.S.A. 107 18533–18538. 10.1073/pnas.1008475107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood M. A., Ibrahim M., Rashid U., Nawaz M., Ali S., Hussain A., et al. (2017). Biomass production for bioenergy using marginal lands. Sustain. Prod. Consum. 9 3–21. 10.1016/j.spc.2016.08.003 [DOI] [Google Scholar]
- Meyfroidt P., Schierhorn F., Prishchepov A. V., Müller D., Kuemmerle T. (2016). Drivers, constraints and trade-offs associated with recultivating abandoned cropland in Russia, Ukraine and Kazakhstan. Glob. Environ. Change 37 1–15. 10.1016/j.gloenvcha.2016.01.003 [DOI] [Google Scholar]
- Mickelbart M. V., Hasegawa P. M., Bailey-Serres J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16 237–251. 10.1038/nrg3901 [DOI] [PubMed] [Google Scholar]
- Mohammed Y. S. A., Tahir I. S. A., Kamal N. M., Eltayeb A. E., Ali A. M., Tsujimoto H. (2014). Impact of wheat-Leymus racemosus added chromosomes on wheat adaptation and tolerance to heat stress. Breed. Sci. 63 450–460. 10.1270/jsbbs.63.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol A. P. (2010). Environmental authorities and biofuel controversies. Environ. Polit. 19 61–79. 10.1080/09644010903396085 [DOI] [Google Scholar]
- Monteith J. (1972). Solar radiation and productivity in tropical ecosystems. J. Appl. Ecol. 9 747–766. [Google Scholar]
- Monteith J. L. (1977). Climate and the efficiency of crop production in Britain. Philos. Trans. R. Soc. Lond. B 281 277–294. 10.1098/rstb.1977.0140 [DOI] [Google Scholar]
- Mortimer J. C., Miles G. P., Brown D. M., Zhang Z., Segura M. P., Weimar T., et al. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. U.S.A. 107 17409–17414. 10.1073/pnas.1005456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K. E., Tilman D., Fornara D. A., Hobbie S. E. (2013). Root depth distribution and the diversity–productivity relationship in a long-term grassland experiment. Ecology 94 787–793. 10.1890/12-1399.1 [DOI] [Google Scholar]
- Mullet J. E. (2017). High-biomass C4 grasses—Filling the yield gap. Plant Sci. 261 10–17. 10.1016/j.plantsci.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Murray S. C., Rooney W. L., Mitchell S. E., Sharma A., Klein P. E., Mullet J. E., et al. (2008). Genetic improvement of sorghum as a biofuel feedstock: II. QTL for stem and leaf structural carbohydrates. Crop Sci. 48 2180–2193. 10.2135/cropsci2008.01.0068 [DOI] [Google Scholar]
- Myburg A., Grattapaglia D., Tuskan G., Jenkins J., Schmutz J., Mizrachi E., et al. (2011). The Eucalyptus grandis Genome Project: genome and transcriptome resources for comparative analysis of woody plant biology. BMC Proc. 5:I20. [Google Scholar]
- Myburg A. A., Grattapaglia D., Tuskan G. A., Hellsten U., Hayes R. D., Grimwood J., et al. (2014). The genome of Eucalyptus grandis. Nature 510 356–362. [DOI] [PubMed] [Google Scholar]
- Nadezhdina N., David T. S., David J. S., Nadezhdin V., Cermak J., Gebauer R., et al. (2012). “Root structure: in situ studies through sap flow research,” inMeasuring Roots - An Updated Approach, Part II Field Methods, ed. Mancuso S. (Heidelberg: Springer; ), 247–266. 10.1007/978-3-642-22067-8_13 [DOI] [Google Scholar]
- Narasimhamoorthy B., Saha M., Swaller T., Bouton J. (2008). Genetic diversity in switchgrass collections assessed by EST-SSR markers. Bioenergy Res. 1 136–146. [Google Scholar]
- National Research Council, (2008). Water Implications of Biofuels Production in the United States. Washington, DC: National Academies Press. [Google Scholar]
- Nijsen M., Smeets E., Stehfest E., Vuuren D. P. (2012). An evaluation of the global potential of bioenergy production on degraded lands. GCB Bioenergy 4 130–147. 10.1111/j.1757-1707.2011.01121.x [DOI] [Google Scholar]
- Nunez-Lopez L., Aguirre-Cruz A., Barrera-Figueroa B. E., Pena-Castro J. M. (2015). Improvement of enzymatic saccharification yield in Arabidopsis thaliana by ectopic expression of the rice SUB1A-1 transcription factor. PeerJ 3:e817. 10.7717/peerj.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, (2017). OECD-FAO Agricultural Outlook 2017-2026. Paris: OECD Publishing. [Google Scholar]
- Orlovic S., Guzina V., Krstic B., Merkulov L. (1998). Genetic variability in anatomical, physiological and growth characteristics of hybrid poplar (Populus x euramericana Dode (Guinier)) and eastern cottonwood (Populus deltoides Bartr.) clones. Silvae Genet. 47 183–189. [Google Scholar]
- Pakull B., Groppe K., Meyer M., Markussen T., Fladung M. (2009). Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet. Genomes 5 505–515. 10.1007/s11295-009-0204-2 [DOI] [Google Scholar]
- Pan Q., Xu Y., Li K., Peng Y., Zhan W., Li W., et al. (2017). The genetic basis of plant architecture in 10 maize recombinant inbred line populations. Plant Physiol. 175 858–873. 10.1104/pp.17.00709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti A., Lambertini C., Monti A. (2018). “Areas with natural constraints to agriculture: possibilities and limitations for the cultivation of switchgrass (Panicum virgatum L.) and Giant Reed (Arundo donax L.) in Europe,” in Land Allocation for Biomass Crops, eds Li R., Monti A., (Berlin: Springer; ), 39–63. 10.1007/978-3-319-74536-7_3 [DOI] [Google Scholar]
- Paterson A. H., Schertz K. F., Lin Y.-R., Liu S.-C., Chang Y.-L. (1995). The weediness of wild plants: molecular analysis of genes influencing dispersal and persistence of johnsongrass, Sorghum halepense (L.) Pers. Proc. Natl. Acad. Sci. U.S.A. 92 6127–6131. 10.1073/pnas.92.13.6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. H., Larson S. R., Keske C. M., Jensen K. B. (2015). “Native grasses for biomass production at high elevations,” inIndustrial Crops Breeding for Bioenergy and Bioproducts, eds Cruz V. M. Z., Dierig D. A., (New York, NY: Springer; ), 101–132. 10.1007/978-1-4939-1447-0_6 [DOI] [Google Scholar]
- Peña-Castro J. M., Van Zanten M., Lee S. C., Patel M. R., Voesenek L. A., Fukao T., et al. (2011). Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 67 434–446. 10.1111/j.1365-313x.2011.04605.x [DOI] [PubMed] [Google Scholar]
- Perez I., Perez J., Carrasco J., Ciria P. (2014). Siberian elm responses to different culture conditions under short rotation forestry in Mediterranean areas. Turk. J. Agric. For. 38 652–662. 10.3906/tar-1311-113 [DOI] [Google Scholar]
- Pérez-Suárez M., Castellano M. J., Kolka R., Asbjornsen H., Helmers M. (2014). Nitrogen and carbon dynamics in prairie vegetation strips across topographical gradients in mixed Central Iowa agroecosystems. Agric. Ecosyst. Environ. 188 1–11. 10.1016/j.agee.2014.01.023 [DOI] [Google Scholar]
- Peterson G. M., Galbraith J. (1932). The concept of marginal land. J. Farm Econ. 14 295–310. [Google Scholar]
- Pierret A., Maeght J.-L., Clément C., Montoroi J.-P., Hartmann C., Gonkhamdee S. (2016). Understanding deep roots and their functions in ecosystems: an advocacy for more unconventional research. Ann. Bot. 118 621–635. 10.1093/aob/mcw130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilu R., Cassani E., Landoni M., Badone F. C., Passera A., Cantaluppi E., et al. (2014). Genetic characterization of an Italian Giant Reed (Arundo donax L.) clones collection: exploiting clonal selection. Euphytica 196 169–181. 10.1007/s10681-013-1022-z [DOI] [Google Scholar]
- Piotrowski S., Carus M., Essel R. (2015). Global bioeconomy in the conflict between biomass supply and demand. Ind. Biotechnol. 11 308–315. 10.1089/ind.2015.29021.stp [DOI] [Google Scholar]
- Pocienë L., Šarûnaitë L., Tilvikienë V., Šlepetys J., Kadžiulienë Ž. (2013). The yield and composition of reed canary grass biomass as raw material for combustion. Biologija 59. [Google Scholar]
- Poovaiah C. R., Mazarei M., Decker S. R., Turner G. B., Sykes R. W., Davis M. F., et al. (2015). Transgenic switchgrass (Panicum virgatum L.) biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol. J. 10 552–563. 10.1002/biot.201400499 [DOI] [PubMed] [Google Scholar]
- Porth I., Klapšte J., Skyba O., Hannemann J., Mckown A. D., Guy R. D., et al. (2013). Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 200 710–726. 10.1111/nph.12422 [DOI] [PubMed] [Google Scholar]
- Portis E., Mauro R. P., Barchi L., Acquadro A., Mauromicale G., Lanteri S. (2014). Mapping yield-associated QTL in globe artichoke. Mol. Breed. 34 615–630. 10.1007/s11032-014-0061-z [DOI] [Google Scholar]
- Portis E., Mauromicale G., Mauro R., Acquadro A., Scaglione D., Lanteri S. (2009). Construction of a reference molecular linkage map of globe artichoke (Cynara cardunculus var. scolymus). Theor. Appl. Genet. 120 59–70. 10.1007/s00122-009-1159-2 [DOI] [PubMed] [Google Scholar]
- Portis E., Scaglione D., Acquadro A., Mauromicale G., Mauro R., Knapp S. J., et al. (2012). Genetic mapping and identification of QTL for earliness in the globe artichoke/cultivated cardoon complex. BMC Res. Notes 5:252. 10.1186/1756-0500-5-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post W. M., Nichols J. A., Wang D., West T. O., Bandaru V., Izaurralde R. C. (2013). Marginal lands: concept, assessment and management. J. Agric. Sci. 5 129–139. [Google Scholar]
- Quinn L. D., Straker K. C., Guo J., Kim S., Thapa S., Kling G., et al. (2015). Stress-tolerant feedstocks for sustainable bioenergy production on marginal land. Bioenergy Res. 8 1081–1100. 10.1007/s12155-014-9557-y [DOI] [Google Scholar]
- Rae A., Robinson K., Street N., Taylor G. (2004). Morphological and physiological traits influencing biomass productivity in short-rotation coppice poplar. Can. J. For. Res. 34 1488–1498. 10.1139/x04-033 [DOI] [Google Scholar]
- Rae A. M., Pinel M. P., Bastien C., Sabatti M., Street N. R., Tucker J., et al. (2008). QTL for yield in bioenergy Populus: identifying G × E interactions from growth at three contrasting sites. Tree Genet. Genomes 4 97–112. [Google Scholar]
- Ranade S. A., Yadav H. (2014). Universal molecular markers for plant breeding and genetics analysis. J. Plant Biochem. Physiol. 2:e121. [Google Scholar]
- Ranalli P. (2004). Current status and future scenarios of hemp breeding. Euphytica 140 121–131. 10.1007/s10681-004-4760-0 [DOI] [Google Scholar]
- Ranjan P., Yin T., Zhang X., Kalluri U. C., Yang X., Jawdy S., et al. (2010). Bioinformatics-based identification of candidate genes from QTLs associated with cell wall traits in Populus. Bioenergy Res. 3 172–182. 10.1007/s12155-009-9060-z [DOI] [Google Scholar]
- Reinhardt D., Kuhlemeier C. (2002). Plant architecture. EMBO Rep. 3 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro B. E. (2013). Beyond commonplace biofuels: social aspects of ethanol. Energy Policy 57 355–362. 10.1016/j.enpol.2013.02.004 [DOI] [Google Scholar]
- Ritter K. B., Jordan D. R., Chapman S. C., Godwin I. D., Mace E. S., Mcintyre C. L. (2008). Identification of QTL for sugar-related traits in a sweet× grain sorghum (Sorghum bicolor L. Moench) recombinant inbred population. Mol. Breed. 22 367–384. 10.1007/s11032-008-9182-6 [DOI] [Google Scholar]
- Robertson B. A., Doran P. J., Loomis E. R., Robertson J. R., Schemske D. W. (2011a). Avian use of perennial biomass feedstocks as post-breeding and migratory stopover habitat. PLoS One 6:e16941. 10.1371/journal.pone.0016941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson B. A., Doran P. J., Loomis L. R., Robertson J. R., Schemske D. W. (2011b). Perennial biomass feedstocks enhance avian diversity. GCB Bioenergy 3 235–246. 10.1111/j.1757-1707.2010.01080.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G. P., Hamilton S. K., Barham B. L., Dale B. E., Izaurralde R. C., Jackson R. D., et al. (2017). Cellulosic biofuel contributions to a sustainable energy future: choices and outcomes. Science 356:eaal2324. 10.1126/science.aal2324 [DOI] [PubMed] [Google Scholar]
- Robertson G. P., Hamilton S. K., Del Grosso S. J., Parton W. J. (2011c). The biogeochemistry of bioenergy landscapes: carbon, nitrogen, and water considerations. Ecol. Appl. 21 1055–1067. 10.1890/09-0456.1 [DOI] [PubMed] [Google Scholar]
- Robson P., Jensen E., Hawkins S., White S. R., Kenobi K., Clifton-Brown J., et al. (2013). Accelerating the domestication of a bioenergy crop: identifying and modelling morphological targets for sustainable yield increase in Miscanthus. J. Exp. Bot. 64 4143–4155. 10.1093/jxb/ert225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson P., Mos M., Clifton-Brown J., Donnison I. (2012). Phenotypic variation in senescence in Miscanthus: towards optimising biomass quality and quantity. Bioenergy Res. 5 95–105. 10.1007/s12155-011-9118-6 [DOI] [Google Scholar]
- Rönnberg-Wästljung A., Glynn C., Weih M. (2005). QTL analyses of drought tolerance and growth for a Salix dasyclados× Salix viminalis hybrid in contrasting water regimes. Theor. Appl. Genet. 110 537–549. 10.1007/s00122-004-1866-7 [DOI] [PubMed] [Google Scholar]
- Ronnberg-Wastljung A., Tsarouhas V., Semirikov V., Lagercrantz U. (2003). A genetic linkage map of a tetraploid Salix viminalis x S. dasyclados hybrid based on AFLP markers. For. Genet. 10 185–194. [Google Scholar]
- Roy S. J., Negrão S., Tester M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26 115–124. 10.1016/j.copbio.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Salentijn E. M., Pereira A., Angenent G. C., Van Der Linden C. G., Krens F., Smulders M. J., et al. (2007). Plant translational genomics: from model species to crops. Mol. Breed. 20 1–13. 10.1007/s11032-006-9069-3 [DOI] [Google Scholar]
- Salentijn E. M., Zhang Q., Amaducci S., Yang M., Trindade L. M. (2015). New developments in fiber hemp (Cannabis sativa L.) breeding. Ind. Crops Prod. 68 32–41. 10.1016/j.indcrop.2014.08.011 [DOI] [Google Scholar]
- Samson R., Girouard P. (1998). Bioenergy opportunities from agriculture. Paper Presented at the Resource Efficient Agricultural Production-Canada, Sainte-Anne-de-Bellevue, 1–4. [Google Scholar]
- Sanghera G. S., Wani S. H., Hussain W., Singh N. (2011). Engineering cold stress tolerance in crop plants. Curr. Genomics 12 30–43. 10.2174/138920211794520178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini A., Fagnani A., Ferrini F., Mittempergher M., Brunetti M., Crivellaro A., et al. (2004). Elm breeding for DED resistance, the Italian clones and their wood properties. For. Syst. 13 179–184. [Google Scholar]
- Sarkar P., Bosneaga E., Auer M. (2009). Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 60 3615–3635. 10.1093/jxb/erp245 [DOI] [PubMed] [Google Scholar]
- Sartoni R., Zegada-Lizarazu W., Monti A. (2015). A new compartmentalised rhizotron system for root phenotyping. Ital. J. Agron. 10 53–58. [Google Scholar]
- Scaglione D., Acquadro A., Portis E., Tirone M., Knapp S. J., Lanteri S. (2012). RAD tag sequencing as a source of SNP markers in Cynara cardunculus L. BMC Genomics 13:3. 10.1186/1471-2164-13-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T., Fernando A. L., Monti A., Rettenmaier N. (2015). Life cycle assessment of bioenergy and bio-based products from perennial grasses cultivated on marginal land in the Mediterranean region. Bioenergy Res. 8 1548–1561. 10.1007/s12155-015-9691-1 [DOI] [Google Scholar]
- Schuler M. L., Mantegazza O., Weber A. P. (2016). Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 87 51–65. [DOI] [PubMed] [Google Scholar]
- Schulze E. D., Körner C., Law B. E., Haberl H., Luyssaert S. (2012). Large-scale bioenergy from additional harvest of forest biomass is neither sustainable nor greenhouse gas neutral. GCB Bioenergy 4 611–616. 10.1111/j.1757-1707.2012.01169.x [DOI] [Google Scholar]
- Scippa G. S., Di Michele M., Di Iorio A., Costa A., Lasserre B., Chiatante D. (2005). The response of Spartium junceum roots to slope: anchorage and gene factors. Ann. Bot. 97 857–866. 10.1093/aob/mcj603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serba D. D., Daverdin G., Bouton J. H., Devos K. M., Brummer E. C., Saha M. C. (2015). Quantitative trait loci (QTL) underlying biomass yield and plant height in switchgrass. Bioenergy Res. 8 307–324. 10.1007/s12155-014-9523-8 [DOI] [Google Scholar]
- Shingu Y., Kudo T., Ohsato S., Kimura M., Ono Y., Yamaguchi I., et al. (2005). Characterization of genes encoding metal tolerance proteins isolated from Nicotiana glauca and Nicotiana tabacum. Biochem. Biophys. Res. Commun. 331 675–680. 10.1016/j.bbrc.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Shortall O. (2013). “Marginal land” for energy crops: Exploring definitions and embedded assumptions. Energy Policy 62 19–27. 10.1016/j.enpol.2013.07.048 [DOI] [Google Scholar]
- Šiaudinis G., Jasinskas A., Šarauskis E., Steponavièius D., Karèauskienë D., Liaudanskienë I. (2015). The assessment of Virginia mallow (Sida hermaphrodita Rusby) and cup plant (Silphium perfoliatum L.) productivity, physico–mechanical properties and energy expenses. Energy 93 606–612. 10.1016/j.energy.2015.09.065 [DOI] [Google Scholar]
- Sixto H., Grau J., Alba N., Alia R. (2005). Response to sodium chloride in different species and clones of genus Populus L. Forestry 78 93–104. 10.1093/forestry/cpi009 [DOI] [Google Scholar]
- Smaliychuk A., Müller D., Prishchepov A. V., Levers C., Kruhlov I., Kuemmerle T. (2016). Recultivation of abandoned agricultural lands in Ukraine: Patterns and drivers. Glob. Environ. Chang. 38 70–81. 10.1016/j.gloenvcha.2016.02.009 [DOI] [Google Scholar]
- Smart L. B., Moskal W. A., Cameron K. D., Bennett A. B. (2001). MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol. 42 686–693. 10.1093/pcp/pce085 [DOI] [PubMed] [Google Scholar]
- Smeets E. M., Faaij A. P., Lewandowski I. M., Turkenburg W. C. (2007). A bottom-up assessment and review of global bio-energy potentials to 2050. Prog. Energy Combust. Sci. 33 56–106. 10.1016/j.pecs.2006.08.001 [DOI] [Google Scholar]
- Soldatos P. (2015). Economic aspects of bioenergy production from perennial grasses in marginal lands of South Europe. Bioenergy Res. 8 1562–1573. 10.1007/s12155-015-9678-y [DOI] [Google Scholar]
- Song Q., Zhang G., Zhu X.-G. (2013). Optimal crop canopy architecture to maximise canopy photosynthetic CO2 uptake under elevated CO2–a theoretical study using a mechanistic model of canopy photosynthesis. Funct. Plant Biol. 40 108–124. [DOI] [PubMed] [Google Scholar]
- Sonnante G., Gatto A., Morgese A., Montemurro F., Sarli G., Blanco E., et al. (2011). Genetic map of artichoke× wild cardoon: toward a consensus map for Cynara cardunculus. Theor. Appl. Genet. 123 1215–1229. 10.1007/s00122-011-1661-1 [DOI] [PubMed] [Google Scholar]
- Swaminathan K., Chae W. B., Mitros T., Varala K., Xie L., Barling A., et al. (2012). A framework genetic map for Miscanthus sinensis from RNAseq-based markers shows recent tetraploidy. BMC Genomics 13:142. 10.1186/1471-2164-13-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder S. K., Saha M. C. (2017). Toward genomics-based breeding in C3 cool-season perennial grasses. Front. Plant Sci. 8:1317. 10.3389/fpls.2017.01317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Liang H., Yan D., Zhao Y., Han X., Carlson J. E., et al. (2013). Populus euphratica: the transcriptomic response to drought stress. Plant Mol. Biol. 83 539–557. 10.1007/s11103-013-0107-3 [DOI] [PubMed] [Google Scholar]
- Teichmann T., Muhr M. (2015). Shaping plant architecture. Front. Plant Sci. 6:233. 10.3389/fpls.2015.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamarus K., Groom K., Murrell J., Byrne M., Moran G. (2002). A genetic linkage map for Eucalyptus globulus with candidate loci for wood, fibre, and floral traits. Theor. Appl. Genet. 104 379–387. 10.1007/s001220100717 [DOI] [PubMed] [Google Scholar]
- Thavamanikumar S., Southerton S., Southeron R., Brawner J., Thumma B. (2018). Eucalypt MAS: Implementation of Marker-Assisted Selection in Australia’s Major Plantation Eucalypts. Melbourne: Gondwana Genomics Pty Ltd. [Google Scholar]
- Thumma B. R., Southerton S. G., Bell J. C., Owen J. V., Henery M. L., Moran G. F. (2010). Quantitative trait locus (QTL) analysis of wood quality traits in Eucalyptus nitens. Tree Genet. Genomes 6 305–317. 10.1007/s11295-009-0250-9 [DOI] [Google Scholar]
- Tian F., Bradbury P. J., Brown P. J., Hung H., Sun Q., Flint-Garcia S., et al. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43 159–162. 10.1038/ng.746 [DOI] [PubMed] [Google Scholar]
- Tilman D., Hill J., Lehman C. (2006). Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 314 1598–1600. 10.1126/science.1133306 [DOI] [PubMed] [Google Scholar]
- Tilman D., Socolow R., Foley J. A., Hill J., Larson E., Lynd L., et al. (2009). Beneficial biofuels—the food, energy, and environment trilemma. Science 325 270–271. 10.1126/science.1177970 [DOI] [PubMed] [Google Scholar]
- Topp C. N., Iyer-Pascuzzi A. S., Anderson J. T., Lee C.-R., Zurek P. R., Symonova O., et al. (2013). 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. U.S.A. 110 E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornqvist C.-E., Taylor M., Jiang Y., Evans J., Buell C. R., Kaeppler S. M., et al. (2018). Quantitative trait locus mapping for flowering time in a Lowland× Upland switchgrass Pseudo-F2 population. Plant Genome 11:170093. 10.3835/plantgenome2017.10.0093 [DOI] [PubMed] [Google Scholar]
- Torres A. F., Noordam-Boot C. M., Dolstra O., Van Der Weijde T., Combes E., Dufour P., et al. (2015a). Cell wall diversity in forage maize: genetic complexity and bioenergy potential. Bioenergy Res. 8 187–202. 10.1007/s12155-014-9507-8 [DOI] [Google Scholar]
- Torres A. F., Van Der Weijde T., Dolstra O., Visser R. G., Trindade L. M. (2013). Effect of maize biomass composition on the optimization of dilute-acid pretreatments and enzymatic saccharification. Bioenergy Res. 6 1038–1051. 10.1007/s12155-013-9337-0 [DOI] [Google Scholar]
- Torres A. F., Visser R. G., Trindade L. M. (2015b). Bioethanol from maize cell walls: genes, molecular tools, and breeding prospects. GCB Bioenergy 7 591–607. 10.1111/gcbb.12164 [DOI] [Google Scholar]
- Trindade L., Dolstra O., Van Loo E. R., Visser R. (2010). Plant breeding and its role in a biobased economy. Biobased Econ. 5 67–82. [Google Scholar]
- Tsarouhas V., Gullberg U., Lagercrantz U. (2002). An AFLP and RFLP linkage map and quantitative trait locus (QTL) analysis of growth traits in Salix. Theor. Appl. Genet. 105 277–288. 10.1007/s00122-002-0918-0 [DOI] [PubMed] [Google Scholar]
- Tsarouhas V., Gullberg U., Lagercrantz U. (2004). Mapping of quantitative trait loci (QTLs) affecting autumn freezing resistance and phenology in Salix. Theor. Appl. Genet. 108 1335–1342. 10.1007/s00122-003-1544-1 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Valentine J., Clifton-Brown J., Hastings A., Robson P., Allison G., Smith P. (2012). Food vs. fuel: the use of land for lignocellulosic ‘next generation’energy crops that minimize competition with primary food production. GCB Bioenergy 4 1–19. 10.1111/j.1757-1707.2011.01111.x [DOI] [Google Scholar]
- Valli F., Trebbi D., Zegada-Lizarazu W., Monti A., Tuberosa R., Salvi S. (2017). In vitro physical mutagenesis of giant reed (Arundo donax L.). GCB Bioenergy 9 1380–1389. [Google Scholar]
- Van Bel M., Diels T., Vancaester E., Kreft L., Botzki A., Van De Peer Y., et al. (2017). PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46 D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weijde T. (2016). Targets and Tools for Optimizing Lignocellulosic Biomass Quality of Miscanthus. Ph.D. thesis, Wageningen University, Wageningen. [Google Scholar]
- van der Weijde T., Alvim Kamei C. L., Torres A. F., Vermerris W., Dolstra O., Visser R. G., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. 10.3389/fpls.2013.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weijde T., Huxley L. M., Hawkins S., Sembiring E. H., Farrar K., Dolstra O., et al. (2017a). Impact of drought stress on growth and quality of miscanthus for biofuel production. GCB Bioenergy 9 770–782. 10.1111/gcbb.12382 [DOI] [Google Scholar]
- van der Weijde T., Kamei C. L. A., Severing E. I., Torres A. F., Gomez L. D., Dolstra O., et al. (2017b). Genetic complexity of miscanthus cell wall composition and biomass quality for biofuels. BMC Genomics 18:406. 10.1186/s12864-017-3802-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weijde T., Kiesel A., Iqbal Y., Muylle H., Dolstra O., Visser R. G., et al. (2017c). Evaluation of Miscanthus sinensis biomass quality as feedstock for conversion into different bioenergy products. GCB Bioenergy 9 176–190. 10.1111/gcbb.12355 [DOI] [Google Scholar]
- Van Esbroeck G., Hussey M., Sanderson M. (1997). Leaf appearance rate and final leaf number of switchgrass cultivars. Crop Sci. 37 864–870. 10.2135/cropsci1997.0011183x003700030028x [DOI] [Google Scholar]
- van Rooijen R., Aarts M. G., Harbinson J. (2015). Natural genetic variation for acclimation of photosynthetic light use efficiency to growth irradiance in Arabidopsis. Plant Physiol. 167 1412–1429. 10.1104/pp.114.252239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbrink J. P., Hilten R. N., Das K., Paterson A. H., Feltus F. A. (2012). Analysis of crystallinity index and hydrolysis rates in the bioenergy crop Sorghum bicolor. Bioenergy Res. 5 387–397. 10.1007/s12155-011-9146-2 [DOI] [Google Scholar]
- Vega K. G., Chavira M. G., De La Vega O. M., Simpson J., Vandemark G. (2001). Analysis of genetic diversity in Agave tequilana var. Azul using RAPD markers. Euphytica 119 335–341. [Google Scholar]
- Verma S. R., Dwivedi U. (2014). Lignin genetic engineering for improvement of wood quality: applications in paper and textile industries, fodder and bioenergy production. S. Afr. J. Bot. 91 107–125. 10.1016/j.sajb.2014.01.002 [DOI] [Google Scholar]
- Vogel J. (2008). Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11 301–307. 10.1016/j.pbi.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Wang D., Lebauer D. S., Dietze M. C. (2010). A quantitative review comparing the yield of switchgrass in monocultures and mixtures in relation to climate and management factors. GCB Bioenergy 2 16–25. 10.1111/j.1757-1707.2010.01035.x [DOI] [Google Scholar]
- Wang G.-J., Nyren P., Xue Q.-W., Aberle E., Eriksmoen E., Tjelde T., et al. (2014). Establishment and yield of perennial grass monocultures and binary mixtures for bioenergy in North Dakota. Agron. J. 106 1605–1613. 10.2134/agronj14.0068 [DOI] [Google Scholar]
- Wang S., Yin Y., Ma Q., Tang X., Hao D., Xu Y. (2012). Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol. 12:138. 10.1186/1471-2229-12-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fan C., Hu H., Li Y., Sun D., Wang Y., et al. (2016). Genetic modification of plant cell walls to enhance biomass yield and biofuel production in bioenergy crops. Biotechnol. Adv. 34 997–1017. 10.1016/j.biotechadv.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2006). Genes controlling plant architecture. Curr. Opin. Biotechnol. 17 123–129. 10.1016/j.copbio.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang M., Liu L., Meng F. (2013). Physiological and proteomic responses of diploid and tetraploid black locust (Robinia pseudoacacia L.) subjected to salt stress. Int. J. Mol. Sci. 14 20299–20325. 10.3390/ijms141020299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling B. P., Dickson T. L., Isaacs R., Gaines H., Gratton C., Gross K. L., et al. (2014). Perennial grasslands enhance biodiversity and multiple ecosystem services in bioenergy landscapes. Proc. Natl. Acad. Sci. U.S.A. 111 1652–1657. 10.1073/pnas.1309492111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling B. P., Meehan T. D., Robertson B. A., Gratton C., Landis D. A. (2011). Biocontrol potential varies with changes in biofuel–crop plant communities and landscape perenniality. GCB Bioenergy 3 347–359. 10.1111/j.1757-1707.2011.01092.x [DOI] [Google Scholar]
- Wu R. (1998). Genetic mapping of QTLs affecting tree growth and architecture in Populus: implication for ideotype breeding. Theor. Appl. Genet. 96 447–457. 10.1007/s001220050761 [DOI] [PubMed] [Google Scholar]
- Wu R., Han Y., Hu J., Fang J., Li L., Li M., et al. (2000). An integrated genetic map of Populus deltoides based on amplified fragment length polymorphisms. Theor. Appl. Genet. 100 1249–1256. 10.1007/s001220051431 [DOI] [Google Scholar]
- Wu X. Y., Kuai B. K., Jia J. Z., Jing H. C. (2012). Regulation of leaf senescence and crop genetic improvement F. J. Integr. Plant Biol. 54 936–952. 10.1111/jipb.12005 [DOI] [PubMed] [Google Scholar]
- Wullschleger S. D. (1993). Biochemical limitations to carbon assimilation in C3 plants—a retrospective analysis of the A/Ci curves from 109 species. J. Exp. Bot. 44 907–920. 10.1093/jxb/44.5.907 [DOI] [Google Scholar]
- Xie G., Peng L. (2011). Genetic engineering of energy crops: a strategy for biofuel production in china free access. J. Integr. Plant Biol. 53 143–150. 10.1111/j.1744-7909.2010.01022.x [DOI] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442 705–708. 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- Xu Z., Zhang D., Hu J., Zhou X., Ye X., Reichel K. L., et al. (2009). Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10:S3. 10.1186/1471-2105-10-S11-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Lewandowski I., Wang X., Yi Z. (2016). Assessment of the production potentials of Miscanthus on marginal land in China. Renew. Sustain. Energy Rev. 54 932–943. 10.1016/j.rser.2015.10.040 [DOI] [Google Scholar]
- Yadav S. K. (2010). Cold stress tolerance mechanisms in plants: a review. Agron. Sustain. Dev. 30 515–527. 10.1051/agro/2009050 [DOI] [Google Scholar]
- Yadav U. P., Ayre B. G., Bush D. R. (2015). Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 6:275. 10.3389/fpls.2015.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Hu Z., Pu Y., Brummer E. C., Ragauskas A. J. (2010). Chemical compositions of four switchgrass populations. Biomass Bioenergy 34 48–53. 10.1016/j.biombioe.2009.09.010 [DOI] [Google Scholar]
- Yang X., Wang X., Ji L., Yi Z., Fu C., Ran J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34 943–958. 10.1007/s00299-015-1756-2 [DOI] [PubMed] [Google Scholar]
- Yin H., Chen C. J., Yang J., Weston D. J., Chen J.-G., Muchero W., et al. (2014). Functional genomics of drought tolerance in bioenergy crops. Crit. Rev. Plant Sci. 33 205–224. 10.1080/07352689.2014.870417 [DOI] [Google Scholar]
- Yin T., Difazio S. P., Gunter L. E., Riemenschneider D., Tuskan G. A. (2004). Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor. Appl. Genet. 109 451–463. 10.1007/s00122-004-1653-5 [DOI] [PubMed] [Google Scholar]
- Zegada-Lizarazu W., Della Rocca G., Centritto M., Parenti A., Monti A. (2018). Giant reed genotypes from temperate and arid environments show different response mechanisms to drought. Physiol. Plant. 163 490–501. 10.1111/ppl.12701 [DOI] [PubMed] [Google Scholar]
- Zegada-Lizarazu W., Elbersen H. W., Cosentino S. L., Zatta A., Alexopoulou E., Monti A. (2010). Agronomic aspects of future energy crops in Europe. Biofuel. Bioprod. Biorefin. 4 674–691. 10.1002/bbb.242 [DOI] [Google Scholar]
- Zeri M., Hussain M. Z., Anderson-Teixeira K. J., Delucia E., Bernacchi C. J. (2013). Water use efficiency of perennial and annual bioenergy crops in central Illinois. J. Geophys. Res. Biogeosci. 118 581–589. 10.1002/jgrg.20052 [DOI] [Google Scholar]
- Zhang B., Tong C., Yin T., Zhang X., Zhuge Q., Huang M., et al. (2009). Detection of quantitative trait loci influencing growth trajectories of adventitious roots in Populus using functional mapping. Tree Genet. Genomes 5 539–552. 10.1007/s11295-009-0207-z [DOI] [Google Scholar]
- Zhang D., Zhang Z., Yang K. (2006). QTL analysis of growth and wood chemical content traits in an interspecific backcross family of white poplar (Populus tomentosa× P. bolleana)× P. tomentosa. Can. J. For. Res. 36 2015–2023. 10.1139/x06-103 [DOI] [Google Scholar]
- Zhang D., Zhang Z., Yang K., Li B. (2004). Genetic mapping in (Populus tomentosa × Populus bolleana) and P. tomentosa Carr. using AFLP markers. Theor. Appl. Genet. 108 657–662. 10.1007/s00122-003-1478-7 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xie M., Tuskan G. A., Muchero W., Chen J.-G. (2018). Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Front. Plant Sci. 9:1535. 10.3389/fpls.2018.01535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. S., Ying-Bai S., Luo X.-F. (2010). Effects of water stress on biomass and photosynthetic characteristics of tetraploid black locust (Robinia pseudoacacia L.), Clones. For. Res. 23 920–923. [Google Scholar]
- Zhao H., Li Q., He J., Yu J., Yang J., Liu C., et al. (2014). Genotypic variation of cell wall composition and its conversion efficiency in Miscanthus sinensis, a potential biomass feedstock crop in China. GCB Bioenergy 6 768–776. 10.1111/gcbb.12115 [DOI] [Google Scholar]
- Zhao T., Holmer R., De Bruijn S., Angenent G. C., Van Den Burg H. A., Schranz M. E. (2017). Phylogenomic synteny network analysis of MADS-box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell 29 1278–1292. 10.1105/tpc.17.00312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Schranz M. E. (2017). Network approaches for plant phylogenomic synteny analysis. Curr. Opin. Plant Biol. 36 129–134. 10.1016/j.pbi.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Zhao X., Yang X., Pei S., He G., Wang X., Tang Q., et al. (2016). The Miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene 586 158–169. 10.1016/j.gene.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang L., Liu D. (2012). Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuel. Bioprod. Biorefin. 6 465–482. 10.1002/bbb.1331 [DOI] [Google Scholar]
- Zhu X.-G., Chang T.-G., Song Q.-F., Finnan J., Barth S., Mårtensson L.-M., et al. (2016). “A Systems approach guiding future biomass crop development on marginal land,” in Perennial Biomass Crops for a Resource-Constrained World, eds Barth S., Murphy-Bokern D., Kalinina O., Taylor G., Jones M. (Berlin: Springer; ), 209–224. 10.1007/978-3-319-44530-4_18 [DOI] [Google Scholar]
- Zhu X.-G., Long S. P., Ort D. R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61 235–261. 10.1146/annurev-arplant-042809-112206 [DOI] [PubMed] [Google Scholar]
- Zuo L.-H., Shang A.-Q., Zhang S., Yu X.-Y., Ren Y.-C., Yang M.-S., et al. (2017). The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS One 12:e0171264. 10.1371/journal.pone.0171264 [DOI] [PMC free article] [PubMed] [Google Scholar]