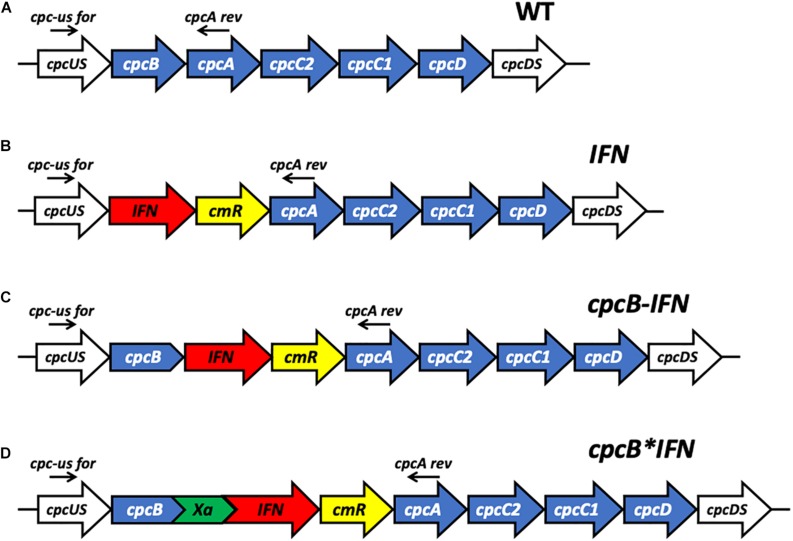
FIGURE 1.
Schematic overview of DNA constructs designed for the transformation of the Synechocystis PCC 6803 (Synechocystis) genome. (A) The native cpc operon, as it occurs in wild type Synechocystis. This DNA sequence and associated strain are referred to as the wild type (WT). (B) Insertion in the cpc operon of the codon-optimized human interferon (IFN) gene followed by the chloramphenicol (cmR) resistance cassette in an operon configuration, replacing the phycocyanin-encoding β-subunit cpcB gene of Synechocystis. This DNA construct is referred to as IFN. (C) Insertion in the cpc operon of the codon-optimized IFN gene immediately downstream of the phycocyanin-encoding β-subunit cpcB gene of Synechocystis, followed by the cmR resistance cassette, in an operon configuration. This DNA construct is referred to as cpcB-IFN. (D) Insertion in the cpc operon of the codon-optimized IFN gene as a fusion construct with the phycocyanin-encoding β-subunit cpcB gene, with the latter in the leader sequence position. The fusion construct cpcB∗IFN was followed by the cmR resistance cassette in an operon configuration. cpcB and IFN genes were linked by the DNA sequence encoding the Factor Xa cleavage site. The latter comprises the Ile-Glu/Asp-Gly-Arg amino acid sequence. This DNA construct is referred to as the cpcB∗IFN.