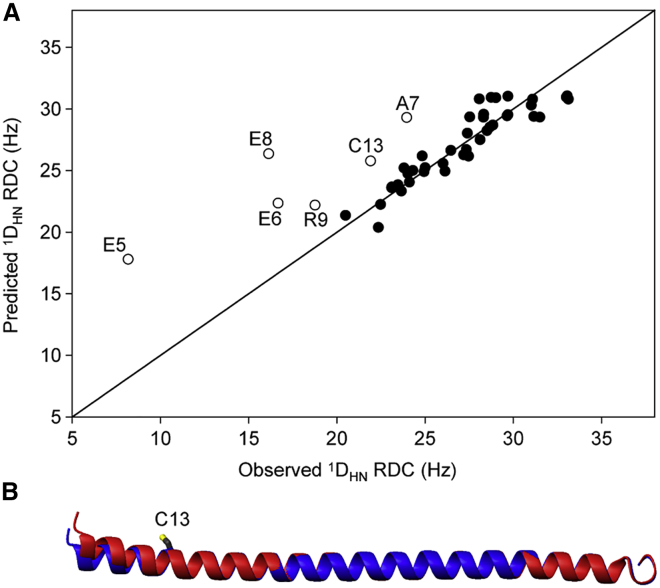
Figure 3.
Effect of I13C mutation on the average structure of the S. scrofa myosin VI SAH domain. (A) Predicted versus observed 1DNH RDCs for the I13C mutant are shown here, using singular value decomposition fitting of the experimental RDCs of residues E15–R64 to the previously determined structure for the WT sequence (Protein Data Bank, PDB: 6OBI) to obtain the alignment tensor. Predicted RDCs for the N-terminal residues are depicted by open symbols. RDCs were measured at 20°C, 900 MHz, in a solution containing ∼11 mg/mL Pf1 and, as mentioned previously (15), were scaled by (R2,max/R2)1/2, where the scaling accounts for the increased dynamics (decreased generalized order parameter, S) when moving toward the termini of the helix. With a Q factor of 9%, the RDCs of residues E15–R64 fit well to those of the 6OBI coordinates. (B) Superposition of the backbone ribbon of the previously determined WT sequence (6OBI, blue) and the I13C mutant (red) is shown, obtained by rigid body refinement, leaving the backbone torsion angles of R12-Q14 harmonically restrained by a weak force constant to their WT values. To see this figure in color, go online.