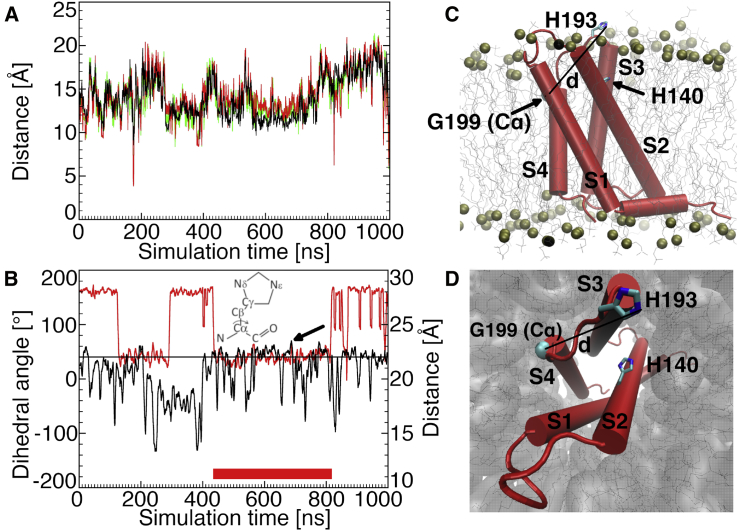
Figure 2.
MD simulation of the monomer without Zn2+ (Mono⋅noZn, MD 1). (A) Time evolution of distances between the H140 and H193 Cα (black), Nδ (green), and Nε atoms (red) is shown. The two histidines are too distant to form one binding site. (B) Time evolution of the distance between the H193 Nε and G199 Cα atoms (black) and of the H140 C-Cα-Cβ-Cγ dihedral angle (red) is shown. Values larger than 22 Å for the distance, above the black line, and lower than 90° for the dihedral angle correlate with the histidine side chains oriented toward the lipid bilayer, between S2 and S3. The thick red line shows the time window during which the side chains of both histidines are most often simultaneously oriented between S2 and S3 (approximately from 400 to 800 ns). The arrow shows the snapshot used for the right panels and for the generation of DimerM via docking. (C) The monomer inserted into the membrane is shown. (D) shows the same as (C), seen from the extracellular bulk solution.