Fig. 1. Arrestin recruitment of N-terminally truncated ACKR3 variants.
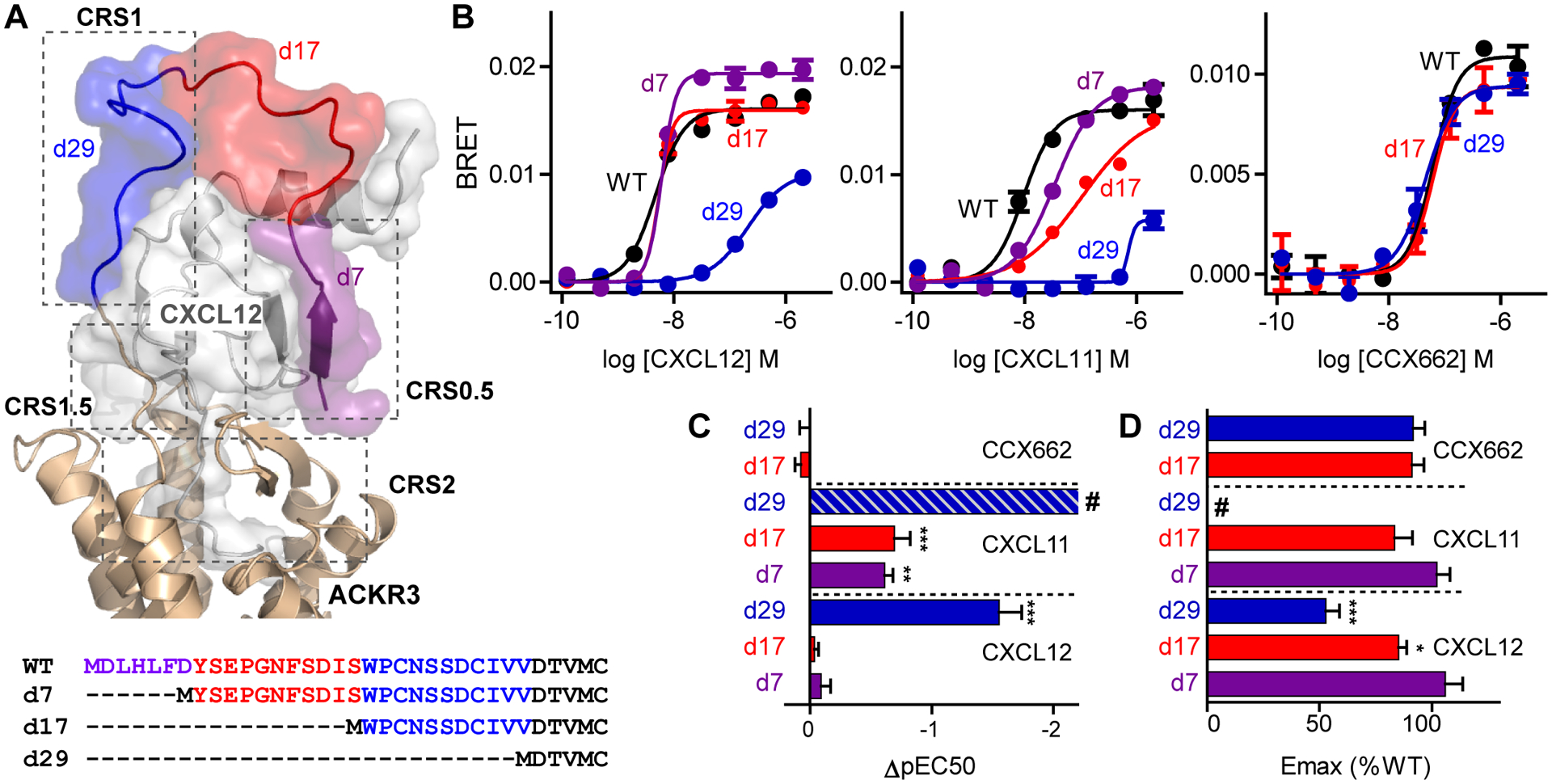
(A) Extracellular portion of the ACKR3:CXCL12 complex from experimentally-driven molecular model (8) and sequences of ACKR3 truncation mutants. Truncated regions are highlighted according to the color scheme in the model. (B) Representative examples of recruitment of GFP10-β-arrestin-2 to ACKR3-Rluc3 variants stimulated with different concentrations of CXCL12WT, CXCL11 and CCX662 measured by BRET. Each point corresponds to the average and standard error of two measurements. (C and D) Potency (C) and efficacy (D) of ligand-induced β-arrestin-2 recruitment determined relative to ACKR3WT from fitting of dose response curves. pEC50 and Emax values for truncation mutants were determined as ΔpEC50=pEC50,mutant-pEC50,WT and %Emax =Emax,mutant/Emax,WT × 100. Data are mean and standard error of three or more experiments. Significantly lowered potency or efficacy relative to ACKR3WT with the same ligand is noted: *P<0.05, **P<0.01, ***P<0.001 from one-way ANOVA with Dunnett’s multiple comparison test. # in (C) indicates that CXCL11-induced arrestin recruitment by ACKR3d29 could not be accurately fit and ΔpEC50 was estimated from the raw data to be >−2.2.
