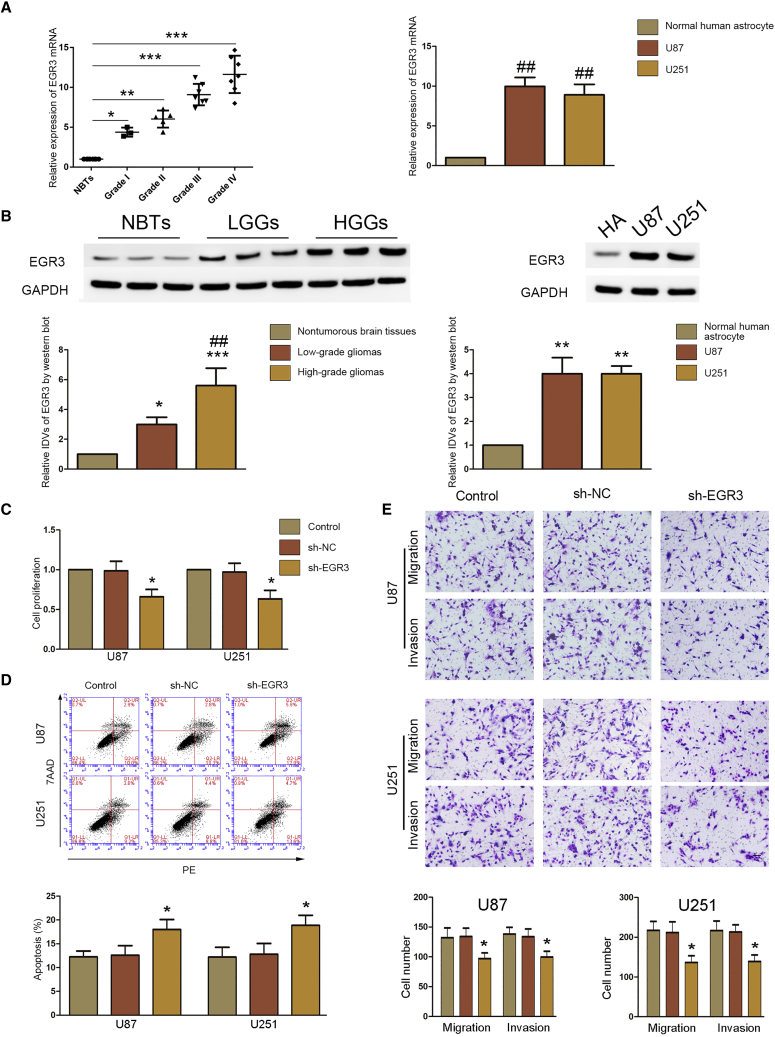
Figure 4.
EGR3 Acts as an Oncogene in Glioma Tissues and Cells
(A) Expression of EGR3 mRNA in glioma tissues (left) and cells (right) was detected by real-time PCR. Data are presented as the mean ± SD (n = 8, NBTs; n = 3, grade I; n = 5, grade II; n = 7, grade III; n = 7, grade IV; n = 3 for each cell line). *p < 0.05 versus NBTs group; **p < 0.01 versus NBTs group; ***p < 0.001 versus NBTs group; ##p < 0.01 versus normal human astrocytes group. (B) western blot was used to detect expression of EGR3 in glioma tissues (left) and cells (right). Data are presented as the mean ± SD (n = 8, NBTs; n = 3, Grade I; n = 5, Grade II; n = 7, Grade III; n = 7, Grade IV; n = 3 each cell line). Left: *p < 0.05 versus nontumorous brain tissues; ***p < 0.001 versus nontumorous brain tissues; ##p < 0.01 versus low-grade glioma tissues. Right: **p < 0.01 versus normal human astrocytes. (C) A CCK-8 assay was conducted to investigate the effect of EGR3 on proliferation in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (D) Flow cytometry analysis of EGR3 in U87 and U251 cells. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. (E) Transwell assays were used to measure the effect of EGR3 on cell migration and invasion in U87 and U251 cells. Representative images and statistical plots are presented. Data are presented as the mean ± SD (n = 3 in each group). *p < 0.05 versus sh-NC group. Scale bar represents 80 μm.