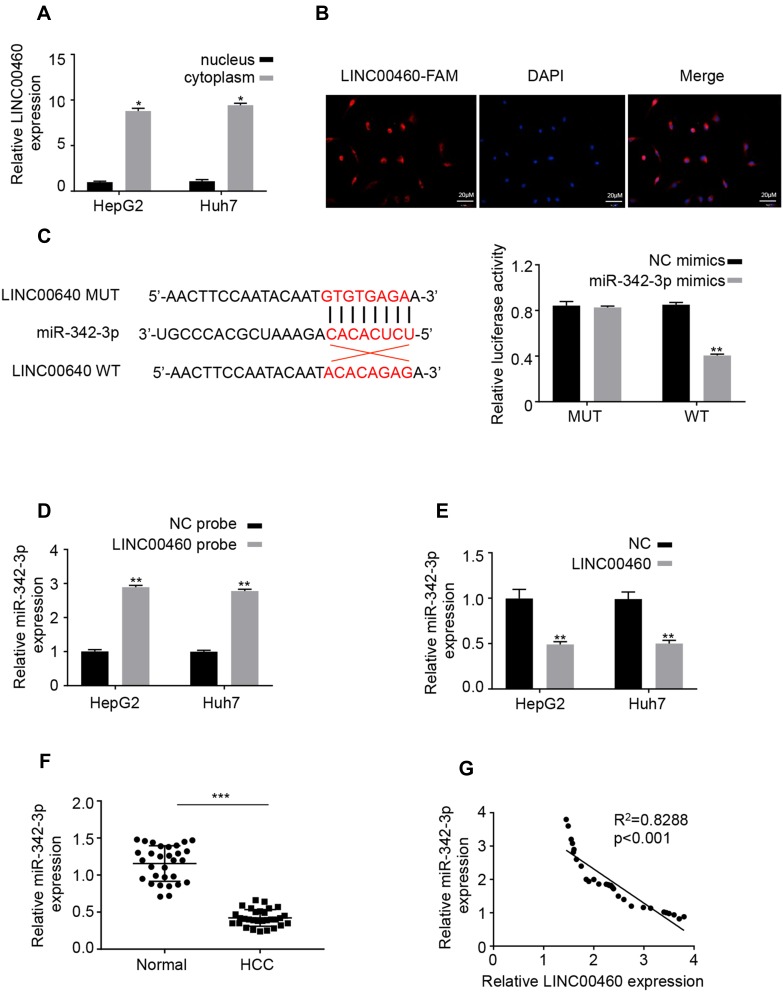
Figure 4.
LINC00460 directly interacts with miR-342-3p in HCC cells. (A) The mRNA levels of nuclear control (U6), cytoplasmic control (GAPDH) and LINC00460 were analyzed using qRT‐PCR in nuclear and cytoplasmic fractions. (B) FISH assay was performed to determine the cellular location of LINC00460. Blue: DAPI (nuclease); Red: LINC00460-FAM (LINC00460). Scale bar, 20 μm. (C). A predicted binding site of miR-342-3p within LINC00460 by bioinformatic analysis using the miRanda software (http://www.microrna.org/). Luciferase activity was determined in HepG2 and Huh7 cells after transfection with miR-342-3p mimic or miRNA negative control (miR-NC). The binding sequences “ACACAGAG” in LINC00460 were mutated to “GTGTGAGA” for generating MUT‐LINC00460. (D and E) RNA pull-down assays were used to determine the interaction between LINC00460 and miR-342-3p. (F) Relative mRNA expression of miR-342-3p in HCC tissues and adjacent normal tissues was assessed using qRT-PCR assay. (G) Pearson correlation analysis between LINC00460 and miR-342-3p expressions in 50 pairs of HCC tissues. All data are representative of three independent experiments and shown as mean ± s.d. *P < 0.05, **P < 0.01, and ***P < 0.001.