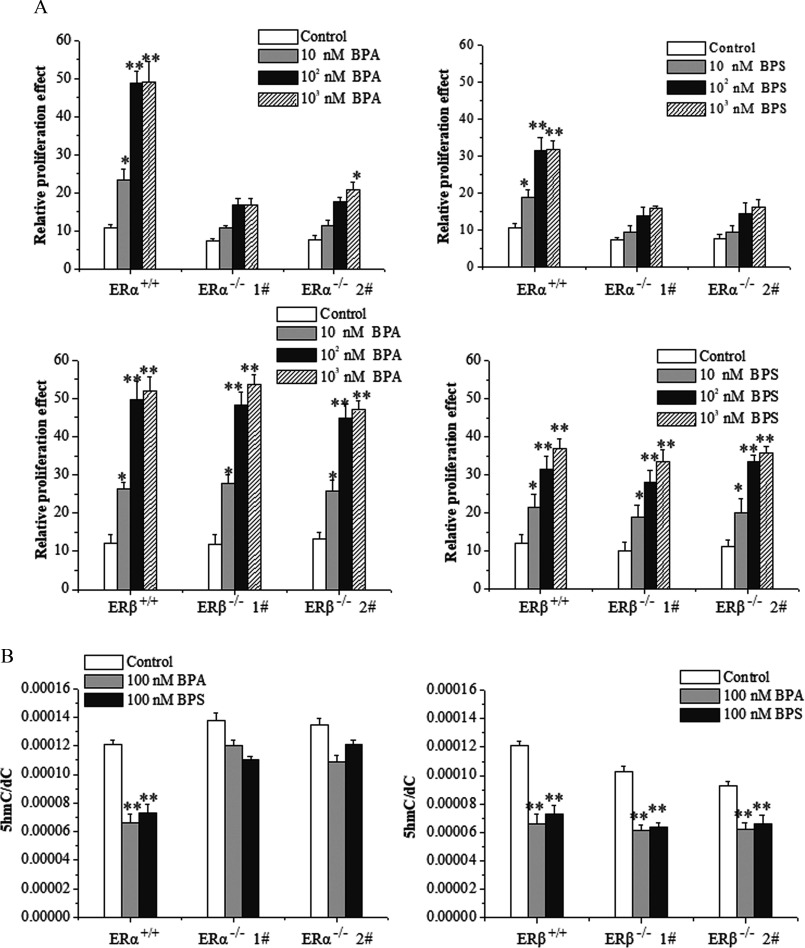
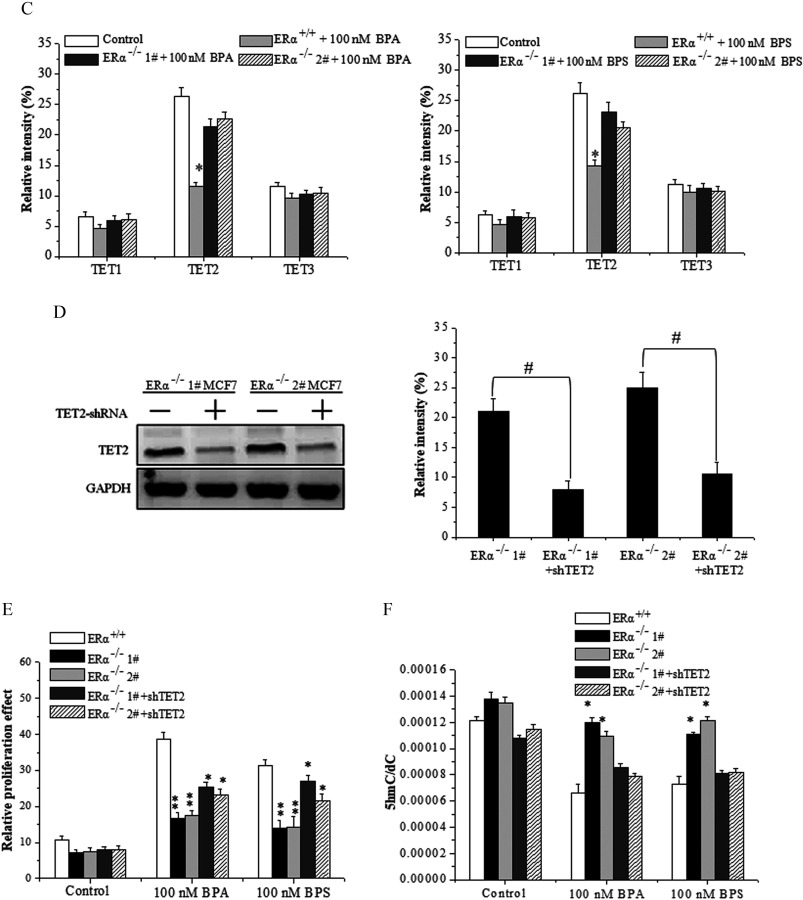
Figure 5.
Cell proliferation, 5-hydroxymethylcytosine () level, and ten-eleven translocation 2 (TET2) expression in estrogen-receptor-knockout MCF-7 cells treated with bisphenol A (BPA) or bisphenol S (BPS). (A) Cell proliferation assay by MTS of and MCF-7 cells exposed to vehicle, BPA, or BPS. Data represent of five independent experiments. Statistical analysis was performed with two-way ANOVA (with Bonferroni posttest): *, or **, vs. control of or cells. (B) frequency measured by UHPLC-MRM MS/MS analysis of digested genomic DNA from and MCF-7 cells exposed to vehicle, BPA, or BPS. Data represent of five independent experiments. Statistical analysis was performed with two-way ANOVA (with Bonferroni posttest). *, or **, vs. control of or cells. (C) Quantification of Western blot detection of TET1, TET2, and TET3 proteins in MCF-7 cells at 48 h after vehicle, BPA, or BPS treatment. The relative intensity was analyzed with ImageJ software (Schneider et al. 2012) and calculated by the ratio relative to the GAPDH intensity. Data represent of three independent experiments. Statistical analysis was performed with Student’s paired t-test. *, vs. control. (D) Identification of TET2 protein in MCF-7 cells transfected with short hairpin RNA (shRNA)-TET2 plasmid using Western blot. The relative intensity was analyzed with ImageJ software and calculated by the ratio relative to the GAPDH intensity. Data represent of three independent experiments. Statistical analysis was performed with Student’s paired t-test. #, vs. indicated samples. (E) Cell proliferation assay by MTS of MCF-7 cells transfected with shRNA-TET2 exposed to BPA and BPS. Data represent of five independent experiments. Statistical analysis was performed with two-way ANOVA (with Bonferroni posttest). *, or **, vs. of each treatment. (F) frequency measured by UHPLC-MRM MS/MS analysis of digested genomic DNA from MCF-7 cells transfected with shRNA-TET2 exposed to BPA and BPS. Data represent of five independent experiments. Statistical significance was evaluated by two-way ANOVA (with Bonferroni posttest). *, or **, vs. control of cells. Note: ANOVA, analysis of variance; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MTS, a tetrazolium salt; SD, standard deviation; UHPLC-MRM MS/MS, ultra-high performance liquid chromatography–tandem mass spectrometry.