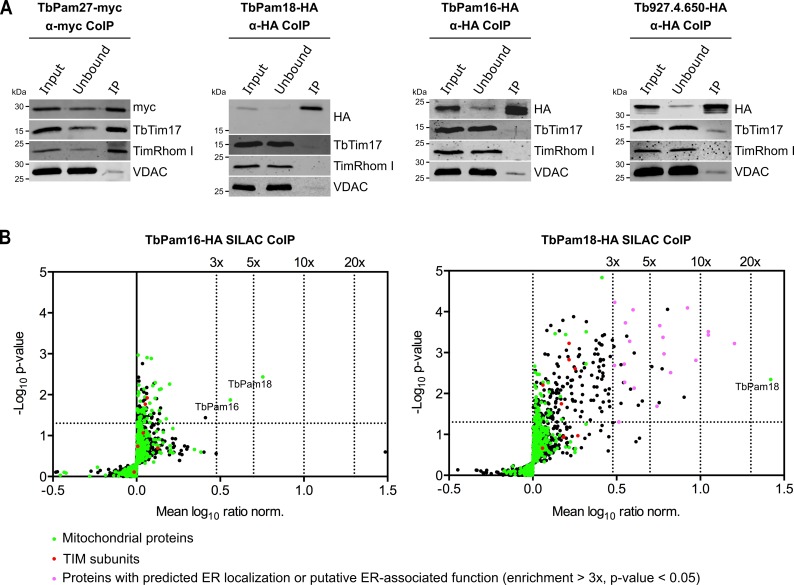
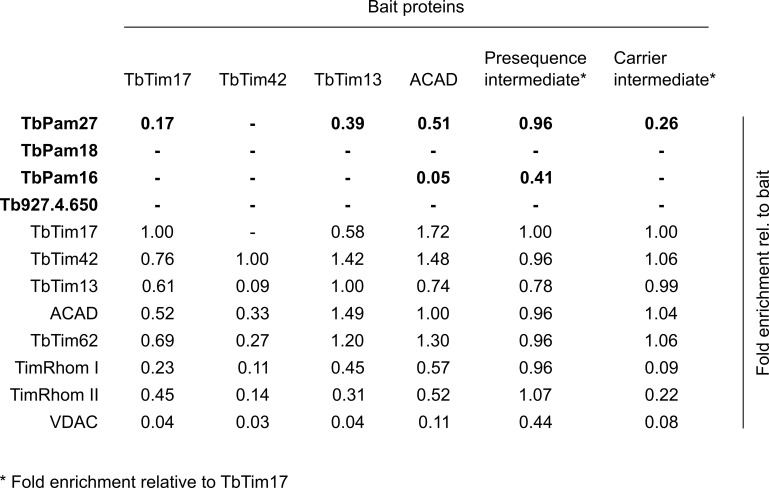
Figure 3. TbPam27, but not TbPam18, TbPam16 and Tb927.4.650, is associated with the TIM complex.
(A) Cell lines expressing the indicated C-terminally triple myc- or HA-tagged proteins were subjected to co-immunoprecipitation (CoIP). 5% each of crude mitochondrial fractions (Input) and unbound proteins (Unbound), as well as 100% of the final eluates (IP) were separated by SDS-PAGE. The resulting immunoblots were probed with anti-tag antibodies and antisera against TIM subunits (TbTim17 and TimRhom I) and VDAC. (B) SILAC-IP experiments of TbPam16-HA and TbPam18-HA from digitonin-extracted mitochondria enriched fractions. Volcano plots show the mean log10 ratios (induced/uninduced) of proteins that were detected in at least two of three independent biological replicates by quantitative MS, plotted against the corresponding -log10 P values (one-sided t-test). The vertical dotted lines specify the indicated enrichment factors. The horizontal dotted line indicates a t-test significance level of 0.05. Green dots represent proteins that are found in the mitochondrial proteome (Peikert et al., 2017), red dots indicate TIM subunits and pink dots represent proteins that are enriched more than three-fold and which, according to the TriTryp database (https://tritrypdb.org/tritrypdb/), are ER-localized or have a putative ER-associated function.