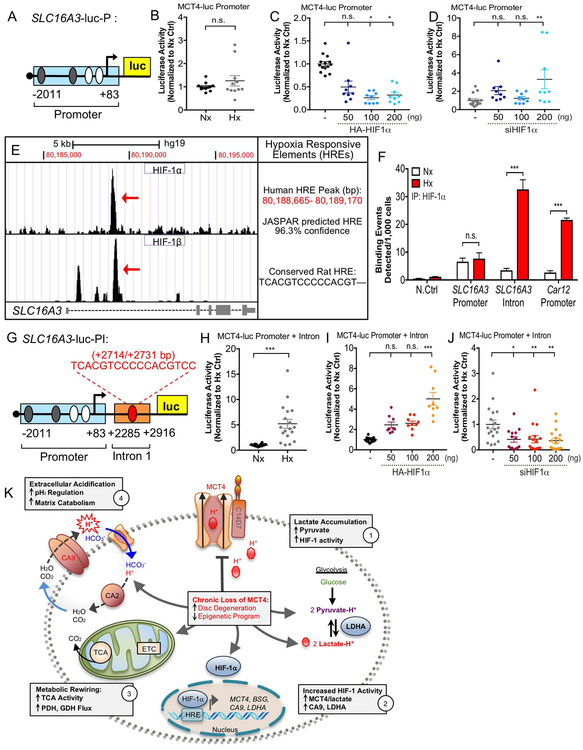
Figure 9: Transcriptional Activation of SLC16A3 in Hypoxia is Controlled by a HIF-1-dependent Intronic Enhancer.
A) Schematic of luciferase reporter plasmid (SLC16A3-luc-P) containing 2kb proximal promoter of SLC16a3 with predicted HREs. HREs in white are previously shown in literature. B-D). SLC16A3-luc-P is not hypoxia-inducible (B) or appropriately HIF-dependent (C, D) in NP cells as determined by dual luciferase assay. E) Analysis of ChIP-sequencing datasets identified novel 500 bp HIF-1 binding region in SLC16a3 intron. JASPAR analysis identified putative HRE site within this region with 96.3% confidence score. Multiz alignment confirmed HRE is conserved in the rat gene. F) Genomic ChIP assay showing HIF-1α binding enrichment along a negative control site, SLC16A3 promoter HRE, SLC16A3 intronic HRE, and positive control HRE on Car12. G) Schematic of SLC16A3-luc-PI plasmid construct with the putative enhancer cloned downstream of the 2 kb proximal promoter. H-J). SLC16A3-luc-PI is hypoxia-inducible (H) and HIF-dependent (I, J) in NP cells. (n=3 independent experiments; 3 technical replicates/experiment). Statistical analysis: t-test, One-way ANOVA. n.s.= not significant; *, p-value ≤ 0.05; **, p-value ≤ 0.01; ***, p-value ≤ 0.001. K) Schematic showing interplay between HIF-1, MCT4 and CA for pH regulation in NP cells. Metabolic consequences of decreased or loss of MCT4 function on NP cell health are depicted.