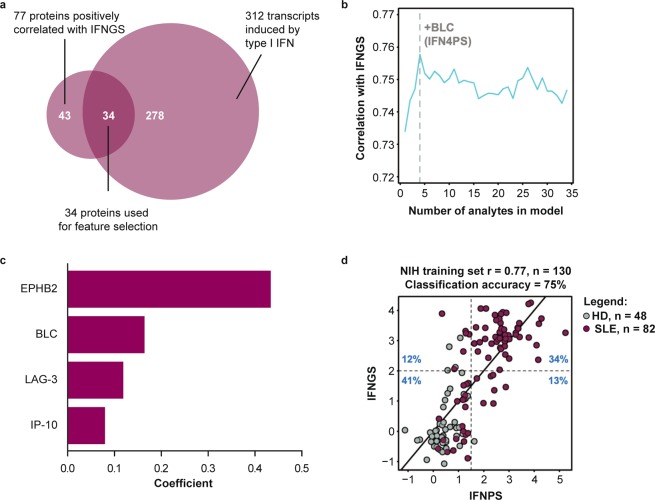
Figure 2.
(a) Venn diagram displaying selection of protein measurements used for feature selection in LASSO regression. 34 SomaLogic protein measurements displayed a Pearson correlation >0.3 versus the interferon gene signature (IFNGS) and were known to have gene expression inducible by type I interferon (IFN) in human cells. Based on the NIH training set (b) average Pearson correlation of IFN protein scores predicted through 10 iterations of 5-fold cross-validation with the IFNGS varying the number of features input into the LASSO regression model. Optimal value of λ was also chosen through 10 iterations of 5-fold cross-validation. Feature selection was restricted to proteins with positive independent associations to the IFNGS. Proteins were scaled to the HD mean and standard deviation prior to fitting the LASSO regression model. (c) Regression coefficients from IFN four-protein signature (IFNPS) refit to IFNGS with ordinary least squares regression. (d) Scatterplot displaying concordance between IFNPS and the IFNGS. r value calculated using Pearson correlation. SLE = systemic lupus erythematosus, HD = healthy donors.