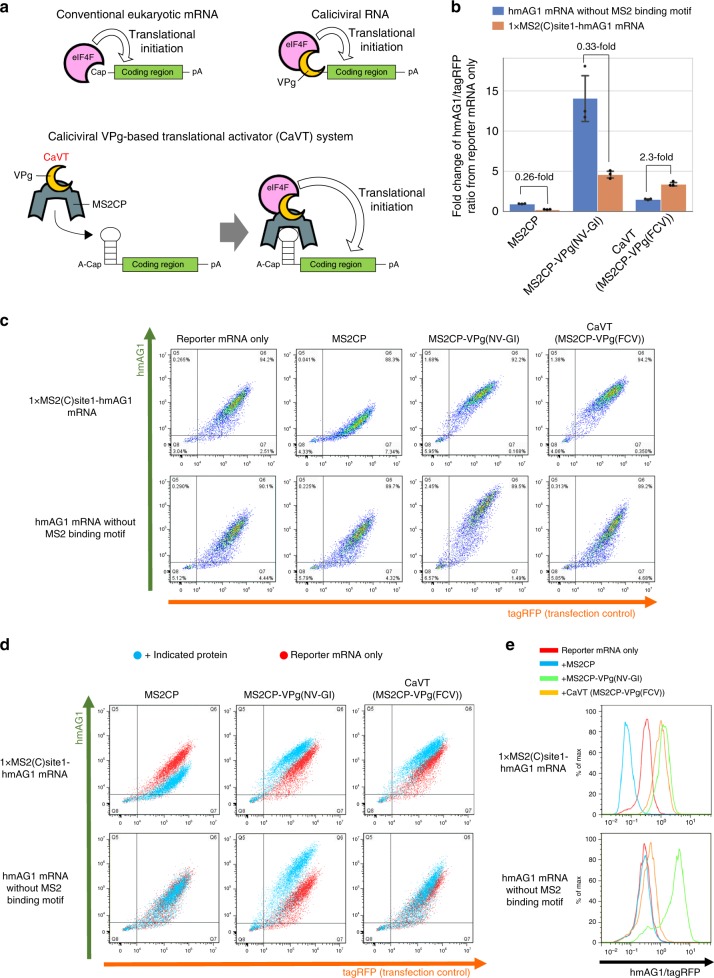
Fig. 2. Comparison of VPg(FCV) and VPg(NV-GI) to construct the translational activator for synthetic mRNAs.
HeLa cells were co-transfected with hmAG1 mRNAs with or without MS2-binding motif (cap analog: A-cap), tagRFP mRNA, and the mRNAs of the indicated proteins. Details of the transfection conditions are described in the Supplementary Methods. The fluorescence was measured by flow cytometry. a Schematic diagrams of the mechanisms of translational initiation. For canonical eukaryotic mRNAs, eIF4F recognizes 5′-cap structures (top left), while for caliciviral RNAs, VPg proteins act as substitutes of the 5′-cap structures (top right). In the case of Caliciviral VPg-based Translational activator (CaVT), MS2CP binds to its target motif at the 5′ UTRs of synthetic mRNAs. eIF4F recognizes the VPg and initiates translation. The basal translation level of these synthetic mRNAs is low because A-cap, a translationally inactive cap analog, is fused with the 5′-end instead of a canonical cap (bottom). b Fold change of the hmAG1/tagRFP ratio caused by each indicated protein. Means of the hmAG1/tagRFP ratio were normalized by the hmAG1/tagRFP ratio in the reporter mRNA only sample. The bar graph shows the average of three independent experiments (mean ± SD). Source data are provided as a Source Data file. c Representative two-dimensional dot plots of hmAG1 and tagRFP. d Superimposition of the dot plots shown in (c). Cells transfected with mRNA to express the indicated proteins are shown as cyan, while cells transfected with only reporter mRNAs are shown as red. e Representative histograms of the hmAG1/tagRFP ratio in cells expressing both hmAG1 and tagRFP.